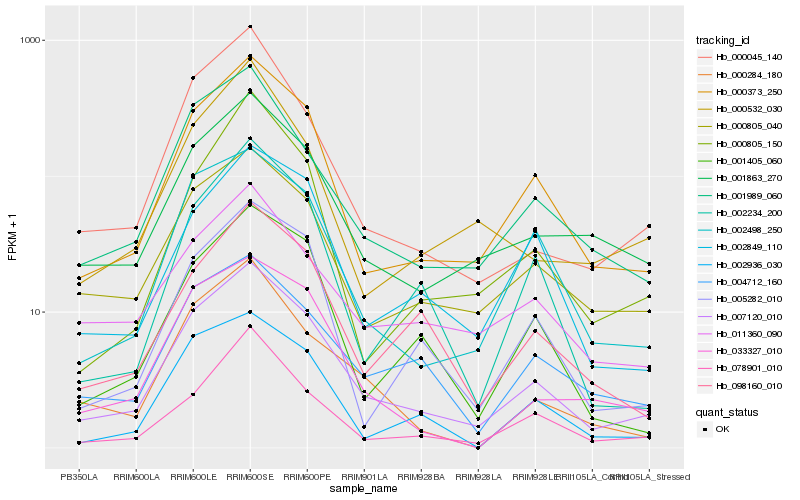
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007120_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |
| 2 |
Hb_000373_250 |
0.0746642997 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 3 |
Hb_001989_060 |
0.1089285413 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000805_040 |
0.1165285801 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 5 |
Hb_001863_270 |
0.1172416389 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 6 |
Hb_000045_140 |
0.1259085753 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 7 |
Hb_002498_250 |
0.1264046785 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_078901_010 |
0.1291480843 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 9 |
Hb_004712_160 |
0.131007277 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 10 |
Hb_033327_010 |
0.1329444829 |
- |
- |
SYM8 [Pisum sativum] |
| 11 |
Hb_001405_060 |
0.1360193396 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 12 |
Hb_005282_010 |
0.1365084888 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 13 |
Hb_098160_010 |
0.1367700824 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000284_180 |
0.1374160797 |
- |
- |
hypothetical protein EUGRSUZ_B01865 [Eucalyptus grandis] |
| 15 |
Hb_002936_030 |
0.1394238065 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002849_110 |
0.1398443834 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 17 |
Hb_002234_200 |
0.1423509812 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 18 |
Hb_000805_150 |
0.142664835 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_011360_090 |
0.1429413824 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 20 |
Hb_000532_030 |
0.1466487862 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |