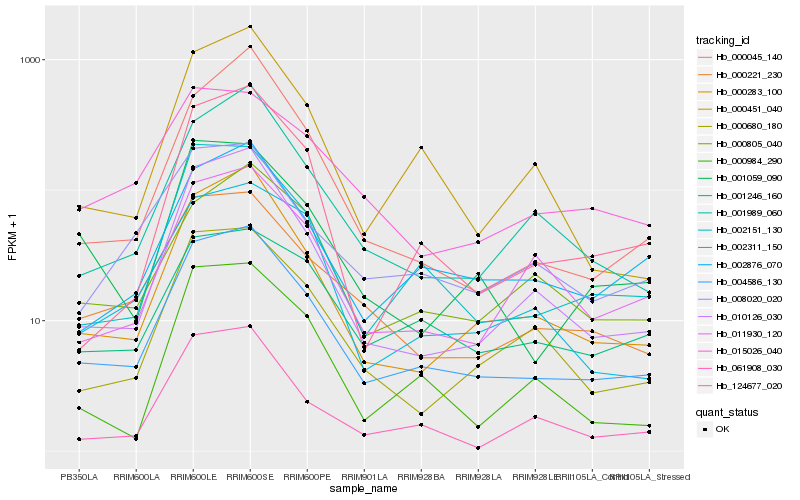
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004586_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000283_100 |
0.0926720881 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002151_130 |
0.098387358 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 4 |
Hb_010126_030 |
0.0999591756 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000221_230 |
0.1048010142 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 6 |
Hb_002876_070 |
0.1148872976 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 7 |
Hb_001989_060 |
0.1165587383 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000680_180 |
0.127833857 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 9 |
Hb_008020_020 |
0.1282338917 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000451_040 |
0.1298977095 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_061908_030 |
0.131046648 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 12 |
Hb_000805_040 |
0.1327854186 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 13 |
Hb_124677_020 |
0.1340258986 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 14 |
Hb_011930_120 |
0.134026915 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002311_150 |
0.1363279618 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 16 |
Hb_001059_090 |
0.1364795157 |
- |
- |
PREDICTED: F-box protein At1g61340 [Jatropha curcas] |
| 17 |
Hb_001246_160 |
0.142577657 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 18 |
Hb_000045_140 |
0.143217366 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 19 |
Hb_000984_290 |
0.1439469669 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_015026_040 |
0.1462559673 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |