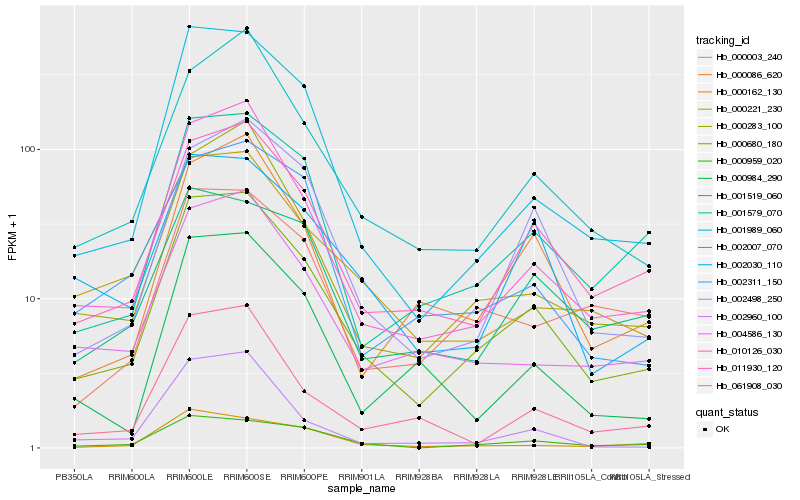
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000680_180 |
0.0 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 2 |
Hb_011930_120 |
0.1001363324 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 3 |
Hb_010126_030 |
0.1009918911 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002007_070 |
0.1048737824 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 5 |
Hb_000283_100 |
0.1186650112 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_001989_060 |
0.1197725412 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001579_070 |
0.123629718 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 8 |
Hb_000959_020 |
0.1267008102 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 9 |
Hb_004586_130 |
0.127833857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002498_250 |
0.129911367 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000221_230 |
0.1363574108 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 12 |
Hb_000086_620 |
0.1371209327 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002030_110 |
0.1434453492 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 14 |
Hb_002311_150 |
0.1445947628 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 15 |
Hb_061908_030 |
0.1454097025 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 16 |
Hb_000003_240 |
0.1458719698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000162_130 |
0.1463346515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000984_290 |
0.1505855452 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001519_060 |
0.1524251353 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_002960_100 |
0.1529088675 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |