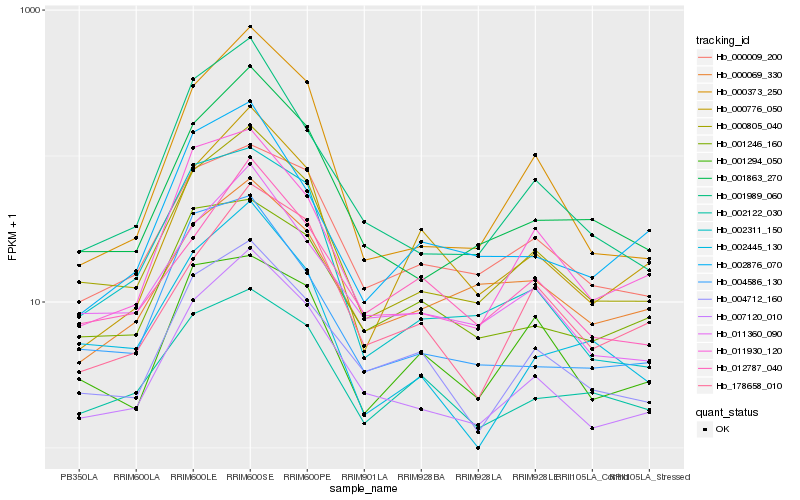
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000805_040 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 2 |
Hb_001863_270 |
0.0808368785 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 3 |
Hb_011360_090 |
0.0916067096 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 4 |
Hb_000009_200 |
0.1113954734 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 5 |
Hb_000373_250 |
0.1164615268 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 6 |
Hb_007120_010 |
0.1165285801 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |
| 7 |
Hb_011930_120 |
0.1165844498 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004712_160 |
0.1184826597 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 9 |
Hb_001989_060 |
0.1197815348 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002122_030 |
0.1217312877 |
- |
- |
- |
| 11 |
Hb_002311_150 |
0.1225625981 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 12 |
Hb_012787_040 |
0.1259816438 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000069_330 |
0.1285189143 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_000776_050 |
0.1298375727 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 15 |
Hb_004586_130 |
0.1327854186 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001246_160 |
0.1330258741 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 17 |
Hb_002876_070 |
0.1414475023 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 18 |
Hb_002445_130 |
0.1425368759 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 19 |
Hb_178658_010 |
0.1428796313 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 20 |
Hb_001294_050 |
0.1443313439 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |