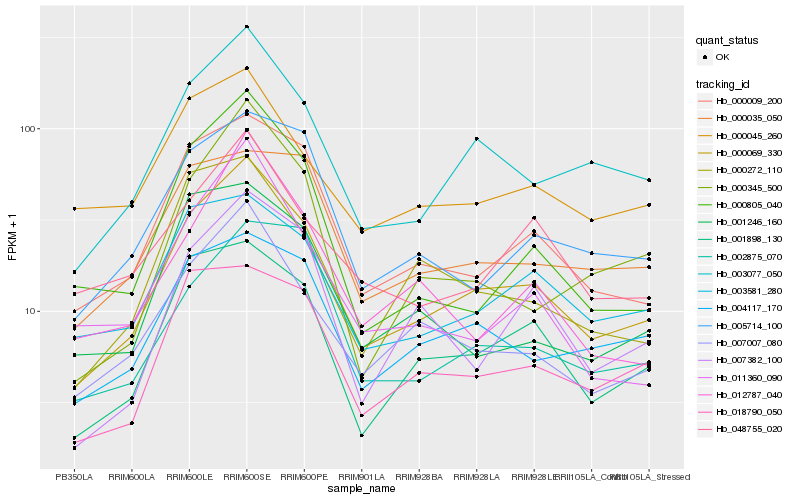
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_330 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_003077_050 |
0.0886675842 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 3 |
Hb_000009_200 |
0.0992617136 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 4 |
Hb_007007_080 |
0.1023660302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001898_130 |
0.1050702725 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_004117_170 |
0.1190370024 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 7 |
Hb_005714_100 |
0.1212709639 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 8 |
Hb_018790_050 |
0.1248182837 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 9 |
Hb_003581_280 |
0.1260577078 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 10 |
Hb_000805_040 |
0.1285189143 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 11 |
Hb_000272_110 |
0.1312057218 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 12 |
Hb_048755_020 |
0.1327358223 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 13 |
Hb_007382_100 |
0.1333847108 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_000045_260 |
0.1350944486 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_000345_500 |
0.1358935117 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 16 |
Hb_000035_050 |
0.1367258144 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 17 |
Hb_011360_090 |
0.1376238399 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 18 |
Hb_012787_040 |
0.1386135596 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002875_070 |
0.1386839203 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 20 |
Hb_001246_160 |
0.1396047927 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |