| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
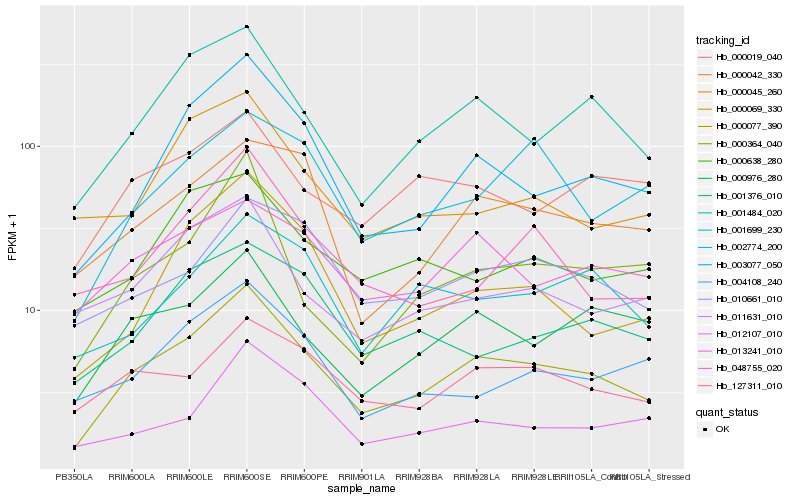
Hb_000077_390 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0019s13330g [Populus trichocarpa] |
| 2 |
Hb_001484_020 |
0.1101557983 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_003077_050 |
0.1207695415 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 4 |
Hb_000976_280 |
0.1328286056 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 5 |
Hb_127311_010 |
0.1418107256 |
- |
- |
hypothetical protein JCGZ_07497 [Jatropha curcas] |
| 6 |
Hb_013241_010 |
0.1438896134 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000042_330 |
0.1455332987 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000069_330 |
0.1468173733 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_010661_010 |
0.1478248302 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 10 |
Hb_011631_010 |
0.1480981887 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 11 |
Hb_001699_230 |
0.1510471572 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 12 |
Hb_004108_240 |
0.1511742732 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 13 |
Hb_001376_010 |
0.1512161923 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 14 |
Hb_000638_280 |
0.1526557252 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 15 |
Hb_048755_020 |
0.1531180586 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 16 |
Hb_000364_040 |
0.1535293321 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF010-like [Jatropha curcas] |
| 17 |
Hb_012107_010 |
0.1538972322 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 18 |
Hb_000019_040 |
0.1541007099 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 19 |
Hb_002774_200 |
0.1542006512 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 20 |
Hb_000045_260 |
0.1597018593 |
- |
- |
protein with unknown function [Ricinus communis] |