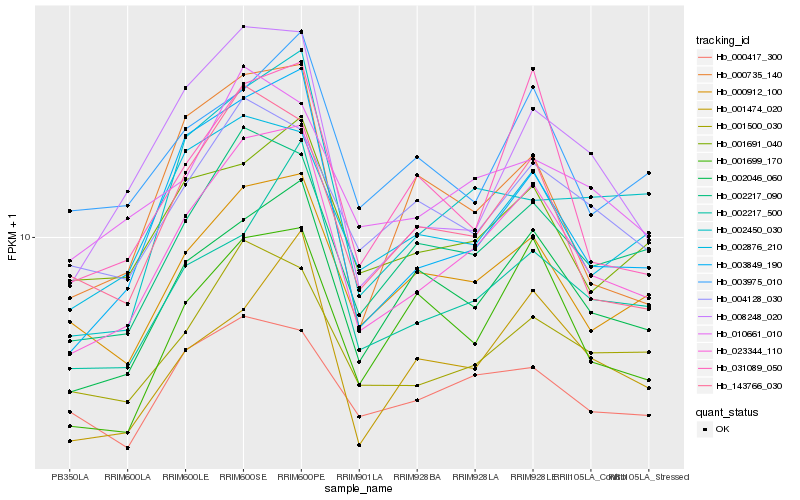
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023344_110 |
0.0 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 2 |
Hb_003849_190 |
0.09938331 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 3 |
Hb_002046_060 |
0.1020668743 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 4 |
Hb_002217_090 |
0.1026248982 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 5 |
Hb_000912_100 |
0.1085286618 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 6 |
Hb_143766_030 |
0.1115125611 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_001500_030 |
0.1116827888 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 8 |
Hb_008248_020 |
0.1170228264 |
transcription factor |
TF Family: DBB |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_002876_210 |
0.1188318848 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 10 |
Hb_002217_500 |
0.1262815323 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 11 |
Hb_001691_040 |
0.1268959061 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |
| 12 |
Hb_031089_050 |
0.1273528967 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_010661_010 |
0.1280614834 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 14 |
Hb_002450_030 |
0.1300624353 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 15 |
Hb_000735_140 |
0.1306061897 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 16 |
Hb_001699_170 |
0.1309781389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004128_030 |
0.1314327932 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 8-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_003975_010 |
0.1322680413 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 19 |
Hb_001474_020 |
0.133218932 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 20 |
Hb_000417_300 |
0.1332843505 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 [Jatropha curcas] |