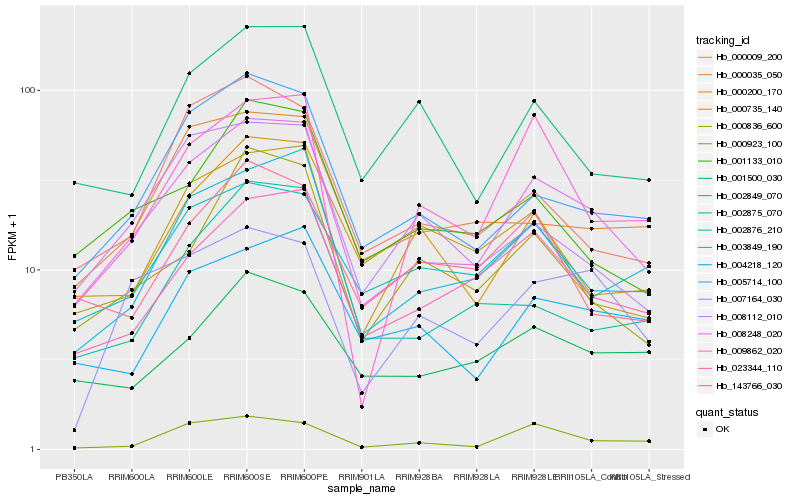
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008248_020 |
0.0 |
transcription factor |
TF Family: DBB |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_003849_190 |
0.0975716005 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 3 |
Hb_023344_110 |
0.1170228264 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 4 |
Hb_005714_100 |
0.1174654984 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 5 |
Hb_000035_050 |
0.1361692074 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 6 |
Hb_001133_010 |
0.1366990682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009862_020 |
0.1390552341 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 8 |
Hb_000836_600 |
0.1417672545 |
- |
- |
PREDICTED: delta(8)-fatty-acid desaturase 2 [Jatropha curcas] |
| 9 |
Hb_004218_120 |
0.142246708 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000735_140 |
0.144567161 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 11 |
Hb_002849_070 |
0.1446106604 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000009_200 |
0.1456509053 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 13 |
Hb_008112_010 |
0.1460792456 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 14 |
Hb_007164_030 |
0.1469511489 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 15 |
Hb_002875_070 |
0.1478589069 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 16 |
Hb_000200_170 |
0.1481064397 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 17 |
Hb_001500_030 |
0.1481797425 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002876_210 |
0.1485768952 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 19 |
Hb_143766_030 |
0.1504063391 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_000923_100 |
0.1505067466 |
- |
- |
chloride channel clc, putative [Ricinus communis] |