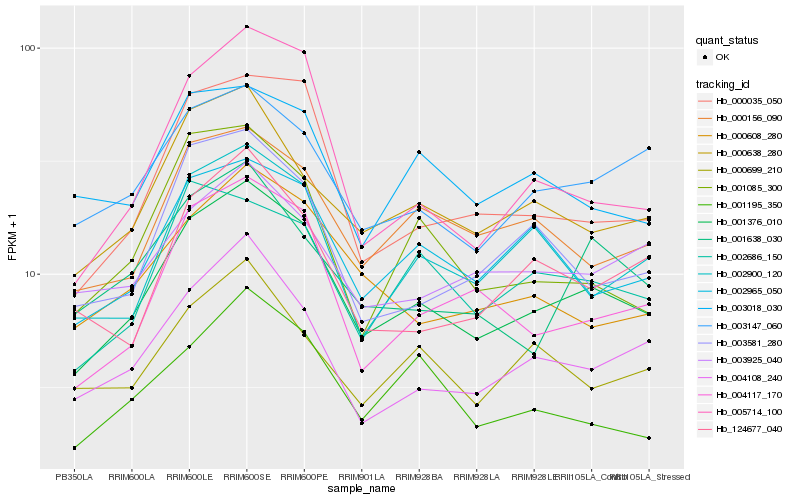
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001376_010 |
0.0 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 2 |
Hb_000035_050 |
0.098166347 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 3 |
Hb_000638_280 |
0.0994517478 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 4 |
Hb_004117_170 |
0.1008863509 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 5 |
Hb_004108_240 |
0.1009408511 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 6 |
Hb_002965_050 |
0.1024261521 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 7 |
Hb_000156_090 |
0.1038420342 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002900_120 |
0.1073994183 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 9 |
Hb_003147_060 |
0.1089487831 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 10 |
Hb_000608_280 |
0.1103541748 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 11 |
Hb_002686_150 |
0.1122273378 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 12 |
Hb_005714_100 |
0.1129973353 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 13 |
Hb_001638_030 |
0.1153983385 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 14 |
Hb_003581_280 |
0.1154166316 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 15 |
Hb_003925_040 |
0.1182166459 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 16 |
Hb_124677_040 |
0.1183703134 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 17 |
Hb_001085_300 |
0.118607948 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 18 |
Hb_000699_210 |
0.1200915545 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 19 |
Hb_003018_030 |
0.1202851394 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 20 |
Hb_001195_350 |
0.1215536636 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |