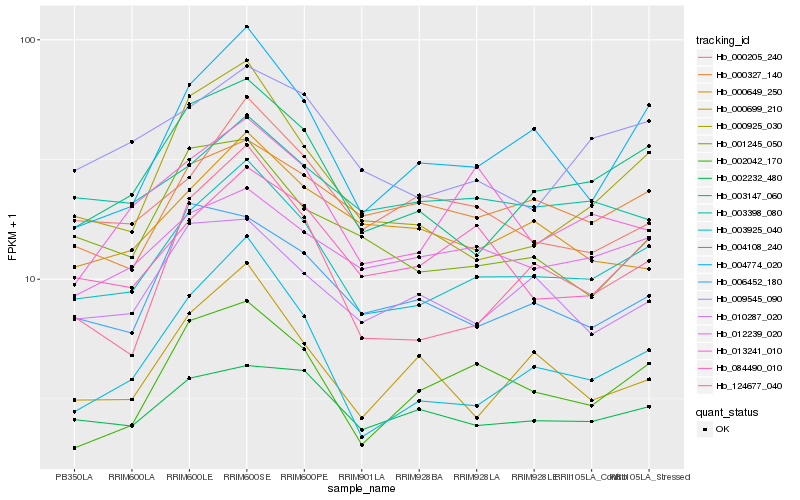
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003925_040 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 2 |
Hb_003147_060 |
0.0772185661 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 3 |
Hb_004774_020 |
0.08650801 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 4 |
Hb_084490_010 |
0.0970074595 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 5 |
Hb_009545_090 |
0.0991569256 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 6 |
Hb_004108_240 |
0.1007609245 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 7 |
Hb_002042_170 |
0.1009764636 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 8 |
Hb_000205_240 |
0.1027234012 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_124677_040 |
0.1040841344 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 10 |
Hb_000925_030 |
0.1043479199 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 11 |
Hb_012239_020 |
0.1051137016 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 12 |
Hb_002232_480 |
0.106206887 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 13 |
Hb_010287_020 |
0.1079642943 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 14 |
Hb_006452_180 |
0.1090365235 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 15 |
Hb_013241_010 |
0.1099072422 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001245_050 |
0.110249202 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 17 |
Hb_003398_080 |
0.1109977601 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000327_140 |
0.1110418018 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000699_210 |
0.1132308659 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 20 |
Hb_000649_250 |
0.1133603139 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |