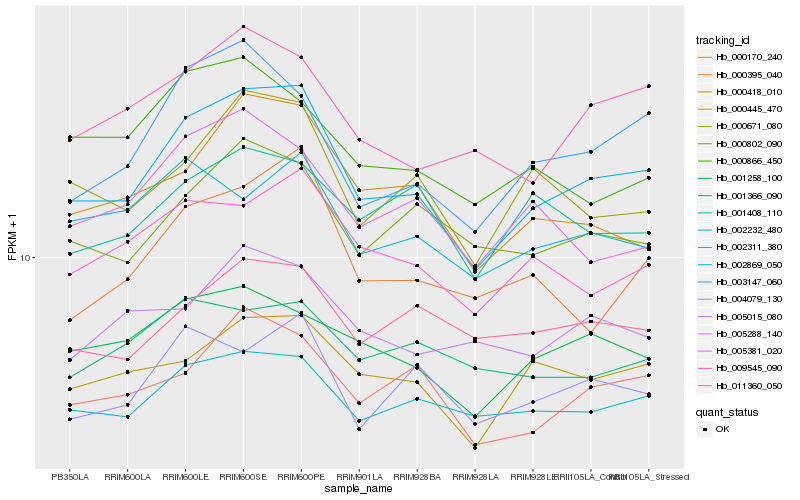
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002869_050 |
0.0 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_002232_480 |
0.090330206 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 3 |
Hb_000170_240 |
0.0951641156 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 4 |
Hb_000445_470 |
0.0953509045 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 1-like [Jatropha curcas] |
| 5 |
Hb_003147_060 |
0.096075521 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 6 |
Hb_005381_020 |
0.0975894972 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 7 |
Hb_009545_090 |
0.0987405516 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 8 |
Hb_000671_080 |
0.0988161535 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 9 |
Hb_000418_010 |
0.0997681442 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 10 |
Hb_000802_090 |
0.101320347 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 11 |
Hb_001258_100 |
0.1014172292 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_004079_130 |
0.1055022932 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 13 |
Hb_000395_040 |
0.1060297802 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 14 |
Hb_011360_050 |
0.1101067519 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005015_080 |
0.1103544329 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005288_140 |
0.1108982764 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 17 |
Hb_002311_380 |
0.1153462625 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 18 |
Hb_000866_450 |
0.116191627 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 19 |
Hb_001366_090 |
0.1168905853 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |
| 20 |
Hb_001408_110 |
0.1178726522 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |