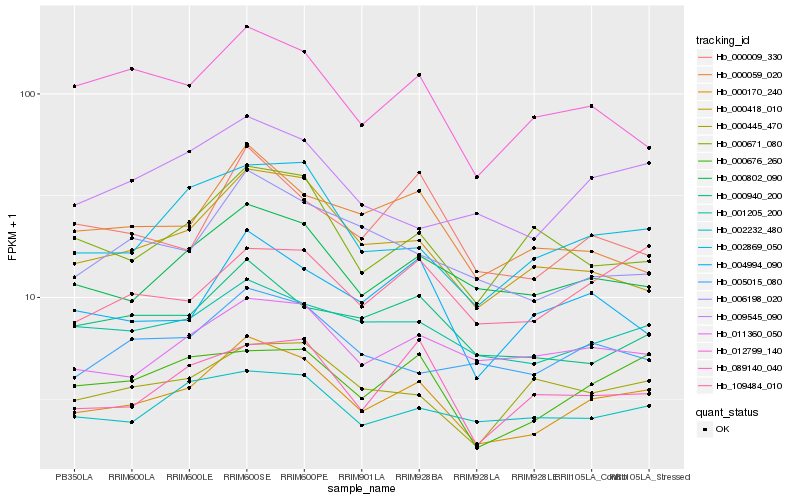
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000170_240 |
0.0 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 2 |
Hb_000802_090 |
0.0917633042 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 3 |
Hb_002869_050 |
0.0951641156 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_001205_200 |
0.1015411006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000676_260 |
0.1016063844 |
- |
- |
PREDICTED: uncharacterized protein LOC105639070 [Jatropha curcas] |
| 6 |
Hb_000418_010 |
0.1057285635 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 7 |
Hb_011360_050 |
0.1074141506 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002232_480 |
0.1120360866 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 9 |
Hb_009545_090 |
0.1124790944 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 10 |
Hb_000940_200 |
0.1128021094 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 11 |
Hb_109484_010 |
0.1131272441 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 12 |
Hb_089140_040 |
0.1132473314 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |
| 13 |
Hb_004994_090 |
0.1139026553 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006198_020 |
0.1144840932 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 15 |
Hb_000009_330 |
0.1150583755 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_000445_470 |
0.1158414597 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 1-like [Jatropha curcas] |
| 17 |
Hb_005015_080 |
0.115992335 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_012799_140 |
0.1169521772 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 19 |
Hb_000671_080 |
0.1171775461 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 20 |
Hb_000059_020 |
0.1172666547 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |