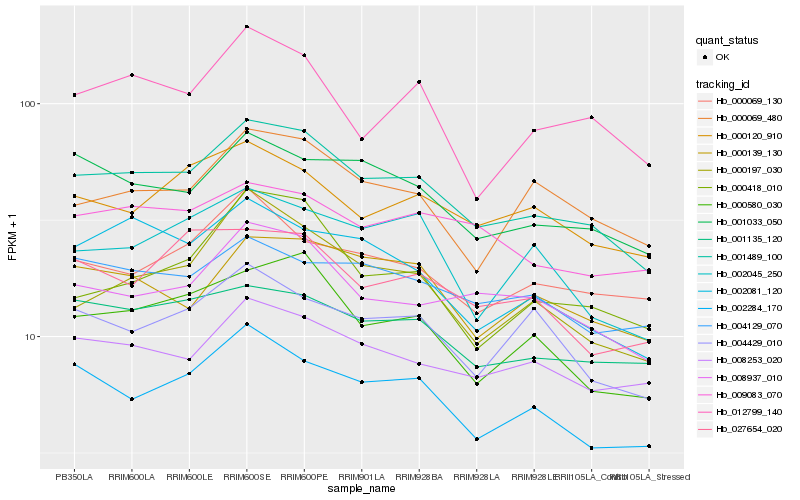
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001489_100 |
0.0 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 2 |
Hb_000580_030 |
0.0660004689 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 3 |
Hb_002284_170 |
0.0689521571 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 4 |
Hb_008937_010 |
0.0690174857 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001033_050 |
0.0697039282 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 6 |
Hb_008253_020 |
0.0736606763 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 7 |
Hb_001135_120 |
0.0739182187 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000139_130 |
0.0762288855 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 9 |
Hb_000120_910 |
0.0788611664 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 10 |
Hb_000069_480 |
0.0791904768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002081_120 |
0.0796434233 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 12 |
Hb_004429_010 |
0.0804495578 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 13 |
Hb_000197_030 |
0.0813117514 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 14 |
Hb_000069_130 |
0.0817986098 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009083_070 |
0.0831020308 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 16 |
Hb_004129_070 |
0.0832517727 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 17 |
Hb_002045_250 |
0.0833832966 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 18 |
Hb_027654_020 |
0.0843425752 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 19 |
Hb_012799_140 |
0.0852712812 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 20 |
Hb_000418_010 |
0.0857868475 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |