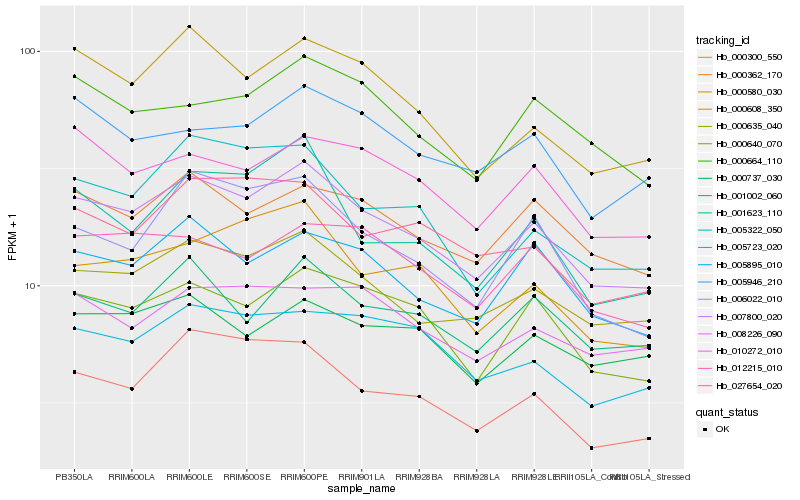
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007800_020 |
0.0 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000635_040 |
0.0535782426 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 3 |
Hb_000640_070 |
0.0629699582 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 4 |
Hb_005895_010 |
0.065820021 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000362_170 |
0.0685829388 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 6 |
Hb_000608_350 |
0.0687401598 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 7 |
Hb_001002_060 |
0.0719601482 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 8 |
Hb_010272_010 |
0.0756718656 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 9 |
Hb_000580_030 |
0.0763662305 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 10 |
Hb_005723_020 |
0.0769844128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000300_550 |
0.077293267 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 12 |
Hb_027654_020 |
0.0773290215 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 13 |
Hb_012215_010 |
0.0775229598 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 14 |
Hb_005322_050 |
0.0785664366 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 15 |
Hb_000664_110 |
0.079403565 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 16 |
Hb_006022_010 |
0.0794095129 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 17 |
Hb_001623_110 |
0.0799784402 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 18 |
Hb_005946_210 |
0.0800986808 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 19 |
Hb_000737_030 |
0.0803258505 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008226_090 |
0.0805671085 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |