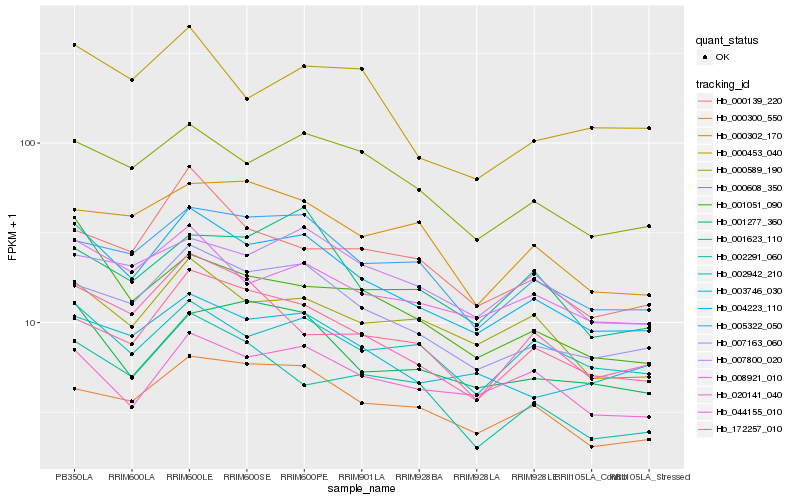
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_110 |
0.0 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 2 |
Hb_007163_060 |
0.0675392299 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 3 |
Hb_000608_350 |
0.0873353969 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 4 |
Hb_008921_010 |
0.0909285682 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |
| 5 |
Hb_172257_010 |
0.0914674967 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 6 |
Hb_000589_190 |
0.0943686007 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005322_050 |
0.0944249636 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 8 |
Hb_003746_030 |
0.0947909744 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 9 |
Hb_000300_550 |
0.0981132466 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 10 |
Hb_001277_360 |
0.1013493133 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 11 |
Hb_020141_040 |
0.106856691 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002942_210 |
0.1074078702 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 13 |
Hb_000453_040 |
0.109980801 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 14 |
Hb_000139_220 |
0.1124292341 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_001051_090 |
0.1128032235 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 16 |
Hb_044155_010 |
0.1142565366 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002291_060 |
0.1149035635 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_007800_020 |
0.1160857563 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001623_110 |
0.1171918729 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_000302_170 |
0.1172948371 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |