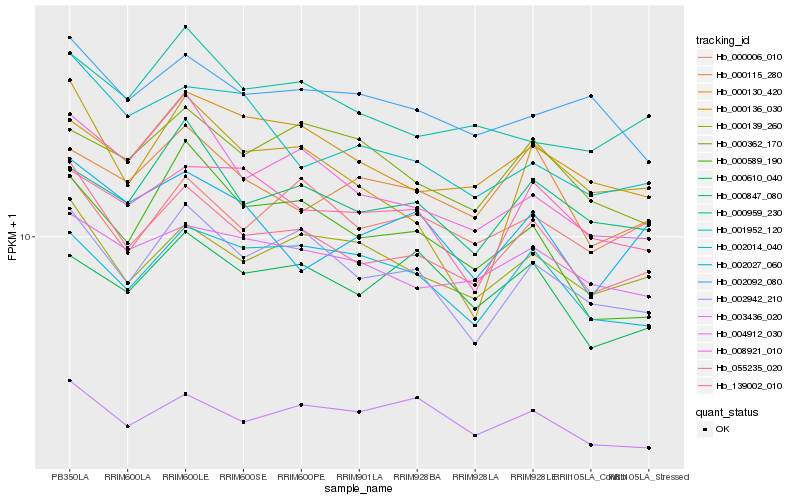
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_139002_010 |
0.0 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_002942_210 |
0.0707428566 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 3 |
Hb_000139_260 |
0.0738006851 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 4 |
Hb_004912_030 |
0.0811969599 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 5 |
Hb_008921_010 |
0.0832911946 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |
| 6 |
Hb_003436_020 |
0.0900279074 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002014_040 |
0.0901924048 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 8 |
Hb_000847_080 |
0.0903658784 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 9 |
Hb_000115_280 |
0.0938899659 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 10 |
Hb_002092_080 |
0.0949658128 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform-like [Jatropha curcas] |
| 11 |
Hb_000130_420 |
0.0952439533 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 12 |
Hb_001952_120 |
0.0952620347 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000006_010 |
0.0952781659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002027_060 |
0.0965498981 |
- |
- |
PREDICTED: exosome complex component RRP4 [Jatropha curcas] |
| 15 |
Hb_055235_020 |
0.0968816763 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000959_230 |
0.0979053766 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 17 |
Hb_000136_030 |
0.0999472498 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 18 |
Hb_000589_190 |
0.101568176 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000610_040 |
0.1017681506 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000362_170 |
0.1022271359 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |