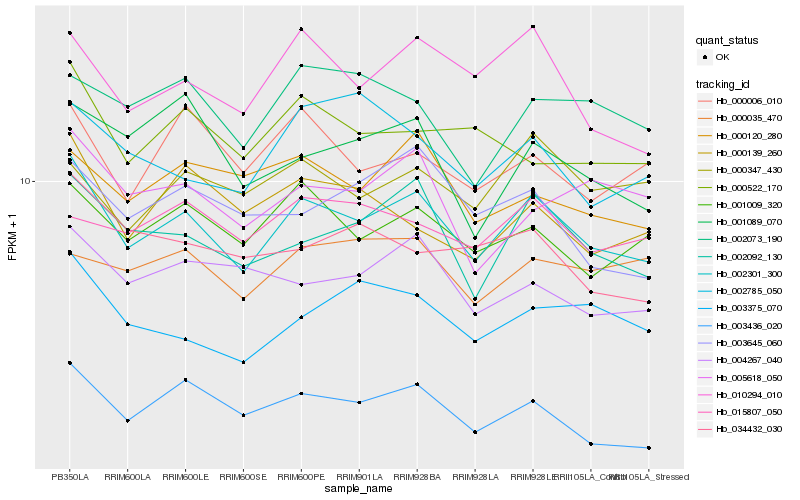
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_300 |
0.0 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 2 |
Hb_002092_130 |
0.0642302668 |
- |
- |
PREDICTED: protein CASP-like [Oryza brachyantha] |
| 3 |
Hb_003436_020 |
0.0700489073 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 4 |
Hb_010294_010 |
0.0704991026 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 5 |
Hb_001009_320 |
0.0725716631 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 6 |
Hb_003375_070 |
0.0800361705 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 7 |
Hb_004267_040 |
0.0827218124 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 8 |
Hb_000522_170 |
0.0832800626 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 9 |
Hb_005618_050 |
0.0833907623 |
- |
- |
PREDICTED: charged multivesicular body protein 7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000120_280 |
0.0839070686 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 11 |
Hb_000006_010 |
0.0842989953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000139_260 |
0.0853759363 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 13 |
Hb_002073_190 |
0.0861433438 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003645_060 |
0.0872539817 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001089_070 |
0.0873842777 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 16 |
Hb_000347_430 |
0.087437329 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 17 |
Hb_002785_050 |
0.0887042932 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 18 |
Hb_034432_030 |
0.089303475 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 19 |
Hb_015807_050 |
0.089443529 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 20 |
Hb_000035_470 |
0.0901594232 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |