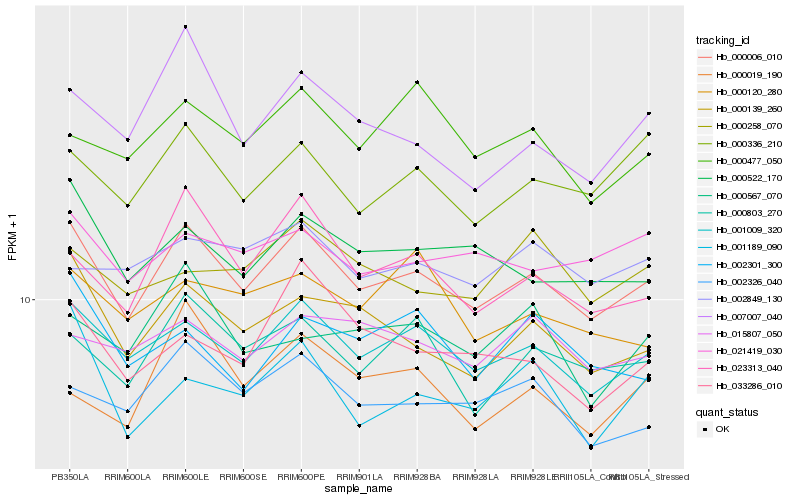
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000006_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001009_320 |
0.0437123884 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 3 |
Hb_000139_260 |
0.0604753643 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 4 |
Hb_023313_040 |
0.0651274643 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000803_270 |
0.0661603048 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 6 |
Hb_000522_170 |
0.0673419451 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 7 |
Hb_000336_210 |
0.0680301041 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 8 |
Hb_002326_040 |
0.0693415727 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 9 |
Hb_007007_040 |
0.073237255 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 10 |
Hb_021419_030 |
0.0748678203 |
- |
- |
hypothetical protein glysoja_023295 [Glycine soja] |
| 11 |
Hb_001189_090 |
0.0799785886 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 12 |
Hb_015807_050 |
0.0809627743 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 13 |
Hb_000477_050 |
0.0811726935 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 14 |
Hb_033286_010 |
0.0812301637 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 15 |
Hb_002301_300 |
0.0842989953 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 16 |
Hb_000120_280 |
0.0845993046 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 17 |
Hb_000258_070 |
0.0849376266 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 18 |
Hb_002849_130 |
0.0851952051 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 19 |
Hb_000567_070 |
0.0852968552 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 20 |
Hb_000019_190 |
0.0857424783 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |