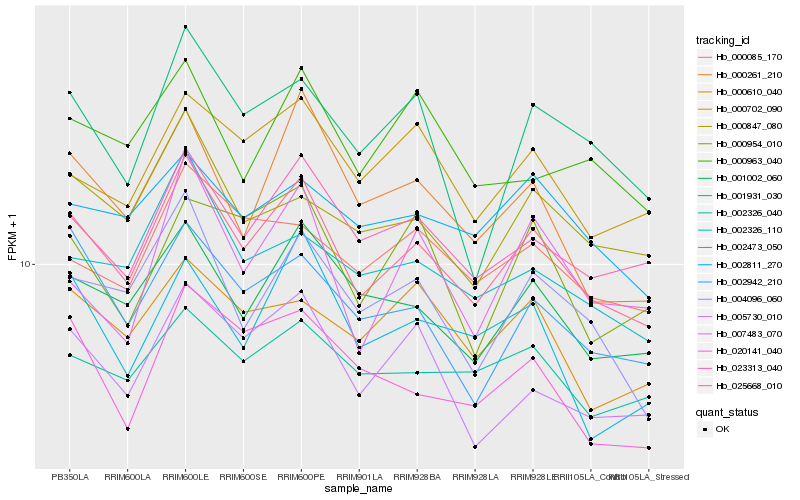
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025668_010 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_023313_040 |
0.0622837162 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001002_060 |
0.0640803849 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 4 |
Hb_004096_060 |
0.0735853204 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 5 |
Hb_005730_010 |
0.0744312142 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 6 |
Hb_002942_210 |
0.0744365508 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 7 |
Hb_000261_210 |
0.0760334188 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 8 |
Hb_000702_090 |
0.0764057909 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 9 |
Hb_000847_080 |
0.0779409973 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 10 |
Hb_002326_110 |
0.0779978861 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 11 |
Hb_001931_030 |
0.0811318521 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta-like [Nelumbo nucifera] |
| 12 |
Hb_002326_040 |
0.0812108407 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 13 |
Hb_000085_170 |
0.0817421888 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_020141_040 |
0.0824009593 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002811_270 |
0.083917753 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 16 |
Hb_000610_040 |
0.0847004398 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002473_050 |
0.0872371458 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_000963_040 |
0.0875114998 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 19 |
Hb_007483_070 |
0.0887897484 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 20 |
Hb_000954_010 |
0.0888990418 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |