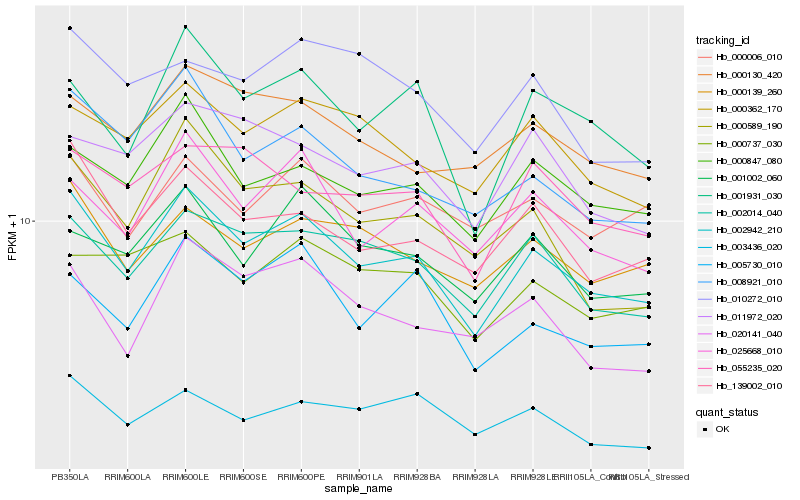
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002942_210 |
0.0 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 2 |
Hb_002014_040 |
0.0690484311 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 3 |
Hb_139002_010 |
0.0707428566 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_008921_010 |
0.0731950669 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |
| 5 |
Hb_025668_010 |
0.0744365508 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000847_080 |
0.0749071053 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 7 |
Hb_001931_030 |
0.0779086869 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta-like [Nelumbo nucifera] |
| 8 |
Hb_000139_260 |
0.0793169412 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 9 |
Hb_000362_170 |
0.0799870214 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 10 |
Hb_005730_010 |
0.0822681865 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 11 |
Hb_001002_060 |
0.0842674846 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 12 |
Hb_000589_190 |
0.0848055463 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000130_420 |
0.0850883969 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 14 |
Hb_020141_040 |
0.0858803866 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000737_030 |
0.0874690883 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003436_020 |
0.087670359 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 17 |
Hb_055235_020 |
0.0898741712 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011972_020 |
0.0899451372 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_000006_010 |
0.0905254495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_010272_010 |
0.0910585247 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |