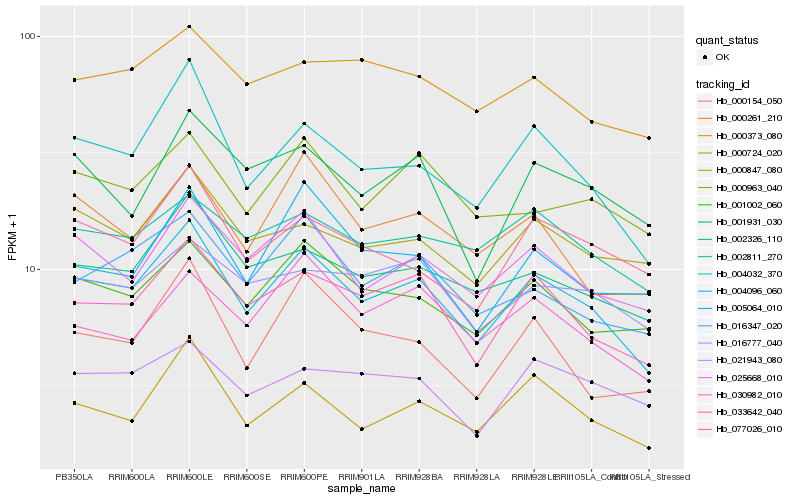
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004096_060 |
0.0 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 2 |
Hb_004032_370 |
0.0603886472 |
- |
- |
pyrophosphate-dependent phosphofructokinase [Hevea brasiliensis] |
| 3 |
Hb_033642_040 |
0.0647132106 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_025668_010 |
0.0735853204 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_000724_020 |
0.0786773662 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 6 |
Hb_001002_060 |
0.0787460436 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 7 |
Hb_002326_110 |
0.0805376499 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 8 |
Hb_016347_020 |
0.084755472 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 9 |
Hb_077026_010 |
0.0904097152 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 10 |
Hb_000847_080 |
0.0908291735 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 11 |
Hb_000261_210 |
0.0910256192 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 12 |
Hb_000963_040 |
0.0912988653 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 13 |
Hb_030982_010 |
0.0920229915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001931_030 |
0.0930688065 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta-like [Nelumbo nucifera] |
| 15 |
Hb_002811_270 |
0.0945689252 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 16 |
Hb_000154_050 |
0.094704496 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 17 |
Hb_016777_040 |
0.095273097 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 18 |
Hb_005064_010 |
0.0955465824 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 19 |
Hb_000373_080 |
0.0969222425 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 20 |
Hb_021943_080 |
0.0969246508 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |