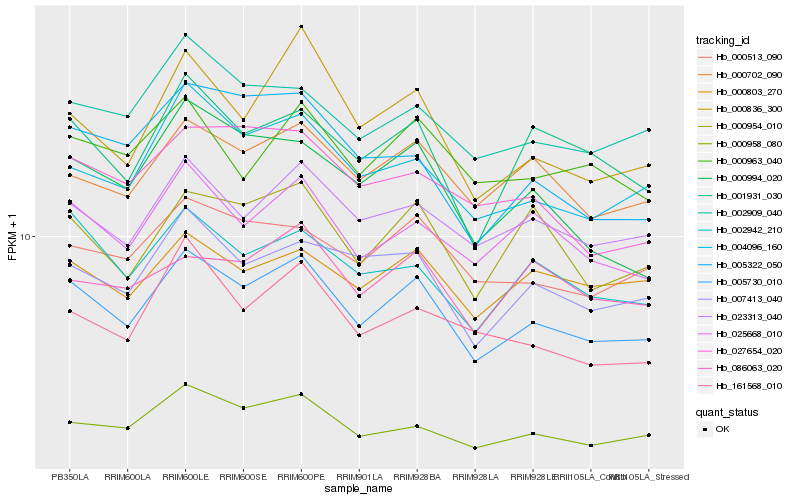
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005730_010 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 2 |
Hb_023313_040 |
0.0743178943 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 3 |
Hb_025668_010 |
0.0744312142 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_000958_080 |
0.0758809519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004096_160 |
0.0760830893 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 6 |
Hb_000702_090 |
0.0764972819 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 7 |
Hb_007413_040 |
0.0768919157 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 8 |
Hb_000954_010 |
0.0800381465 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 9 |
Hb_001931_030 |
0.0810830976 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta-like [Nelumbo nucifera] |
| 10 |
Hb_000513_090 |
0.0817589592 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 11 |
Hb_002942_210 |
0.0822681865 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 12 |
Hb_000836_300 |
0.084136048 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 13 |
Hb_002909_040 |
0.0845989254 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 14 |
Hb_000994_020 |
0.0869633876 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000963_040 |
0.0886475403 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 16 |
Hb_000803_270 |
0.0887916928 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 17 |
Hb_027654_020 |
0.0920818672 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 18 |
Hb_161568_010 |
0.0925446418 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 19 |
Hb_005322_050 |
0.0927811279 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 20 |
Hb_086063_020 |
0.0947244235 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |