| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
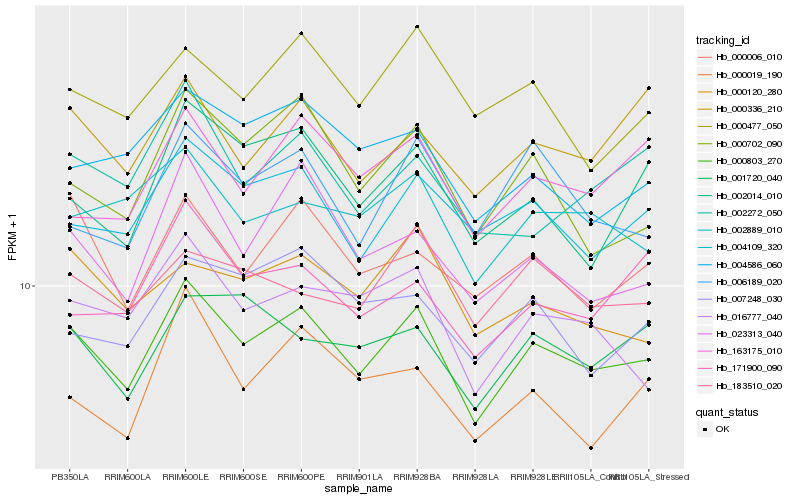
Hb_000803_270 |
0.0 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 2 |
Hb_004109_320 |
0.0616016496 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 3 |
Hb_023313_040 |
0.0649458071 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000006_010 |
0.0661603048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002889_010 |
0.0675050364 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 6 |
Hb_000702_090 |
0.0681936574 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 7 |
Hb_183510_020 |
0.0690183447 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 8 |
Hb_000336_210 |
0.0690598772 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000120_280 |
0.0691768396 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 10 |
Hb_002014_010 |
0.0692425407 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_016777_040 |
0.0692734948 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 12 |
Hb_001720_040 |
0.0698168428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000477_050 |
0.0700518426 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 14 |
Hb_163175_010 |
0.0708806313 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 15 |
Hb_004586_060 |
0.0711306968 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 16 |
Hb_006189_020 |
0.0712839719 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 17 |
Hb_002272_050 |
0.0721923771 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 18 |
Hb_000019_190 |
0.0723741124 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 19 |
Hb_171900_090 |
0.0728305875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007248_030 |
0.0738606366 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |