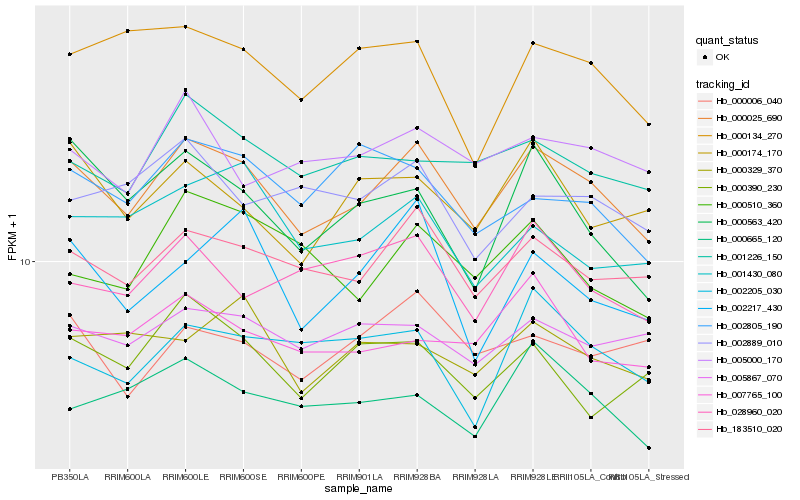
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_690 |
0.0 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 2 |
Hb_183510_020 |
0.0572487597 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 3 |
Hb_001226_150 |
0.0753837046 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 4 |
Hb_005867_070 |
0.0764967715 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000665_120 |
0.0772437148 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 6 |
Hb_000174_170 |
0.0776768368 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007765_100 |
0.0799500317 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 8 |
Hb_028960_020 |
0.0804406091 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000006_040 |
0.0806686827 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 10 |
Hb_000134_270 |
0.0811018634 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 11 |
Hb_005000_170 |
0.0811747768 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 12 |
Hb_000563_420 |
0.0812222071 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 13 |
Hb_000329_370 |
0.0819407093 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 14 |
Hb_000510_360 |
0.0820747833 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 15 |
Hb_001430_080 |
0.0825056824 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 16 |
Hb_002805_190 |
0.084137242 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 17 |
Hb_002205_030 |
0.0841728529 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 18 |
Hb_002217_430 |
0.0846997392 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 19 |
Hb_000390_230 |
0.0849287037 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 20 |
Hb_002889_010 |
0.0851349452 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |