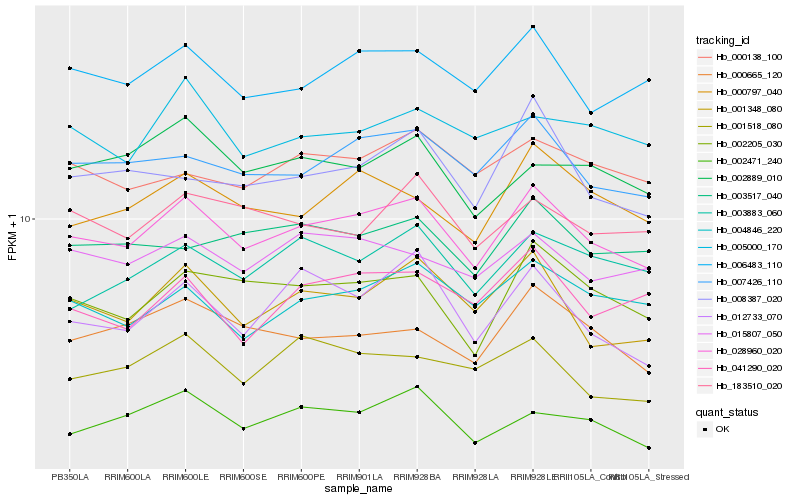
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028960_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007426_110 |
0.052518019 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 3 |
Hb_006483_110 |
0.0557182256 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 4 |
Hb_001348_080 |
0.0567629698 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 5 |
Hb_002205_030 |
0.0581327755 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 6 |
Hb_000797_040 |
0.0622359206 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 7 |
Hb_004846_220 |
0.0629139259 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000665_120 |
0.0642491232 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 9 |
Hb_015807_050 |
0.0661050486 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 10 |
Hb_002471_240 |
0.0666742163 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008387_020 |
0.0682073264 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 12 |
Hb_003517_040 |
0.0688441402 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003883_060 |
0.0696533151 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000138_100 |
0.0700227932 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 15 |
Hb_183510_020 |
0.0702899854 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 16 |
Hb_041290_020 |
0.0705752135 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 17 |
Hb_005000_170 |
0.071327903 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 18 |
Hb_012733_070 |
0.0715262352 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 19 |
Hb_002889_010 |
0.0715829279 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 20 |
Hb_001518_080 |
0.0718963863 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |