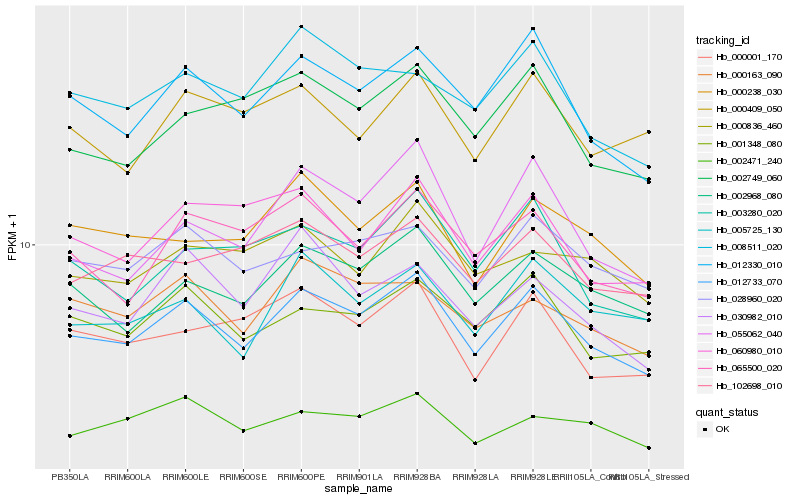
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012733_070 |
0.0 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 2 |
Hb_012330_010 |
0.056071678 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_002968_080 |
0.0621385944 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 4 |
Hb_001348_080 |
0.0655822122 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 5 |
Hb_028960_020 |
0.0715262352 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003280_020 |
0.0733804384 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_065500_020 |
0.0738337295 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 8 |
Hb_005725_130 |
0.0763899091 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 9 |
Hb_002749_060 |
0.0764575903 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 10 |
Hb_000001_170 |
0.0767334936 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_002471_240 |
0.077067482 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000238_030 |
0.0772690356 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_000163_090 |
0.0780684535 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 14 |
Hb_055062_040 |
0.0790683216 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 15 |
Hb_008511_020 |
0.0799287314 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 16 |
Hb_102698_010 |
0.0808034713 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_060980_010 |
0.0809086527 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 18 |
Hb_000409_050 |
0.0823279407 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 19 |
Hb_030982_010 |
0.0830984409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000836_460 |
0.0839134164 |
- |
- |
PREDICTED: uncharacterized protein LOC105628435 [Jatropha curcas] |