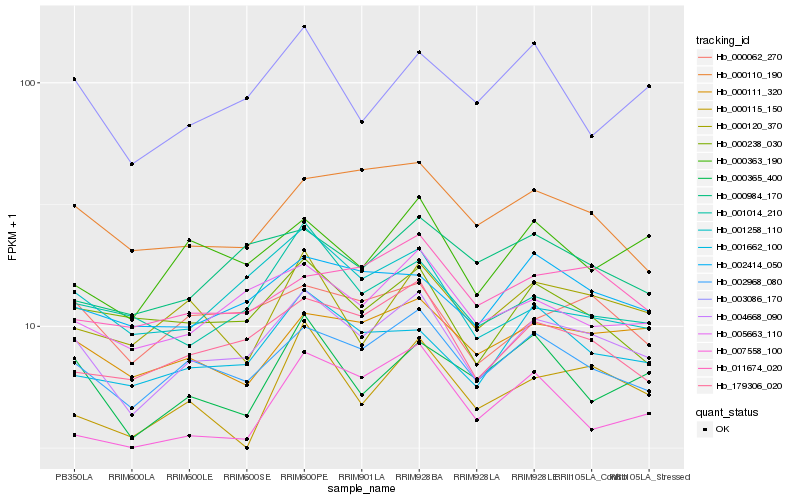
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004668_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630440 [Jatropha curcas] |
| 2 |
Hb_002968_080 |
0.0606090329 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 3 |
Hb_179306_020 |
0.0786745675 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005663_110 |
0.0806545912 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000062_270 |
0.0817650661 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001662_100 |
0.0849957154 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 7 |
Hb_000111_320 |
0.0851229363 |
transcription factor |
TF Family: bZIP |
Transcription factor HBP-1b(c1), putative [Ricinus communis] |
| 8 |
Hb_002414_050 |
0.0861944683 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 9 |
Hb_000363_190 |
0.0913081811 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000238_030 |
0.0919874166 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_001258_110 |
0.0933397215 |
- |
- |
transporter, putative [Ricinus communis] |
| 12 |
Hb_000115_150 |
0.0942292867 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 13 |
Hb_000365_400 |
0.0948937632 |
- |
- |
metal ion transporter, putative [Ricinus communis] |
| 14 |
Hb_011674_020 |
0.0956845916 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 regulatory subunit-like [Jatropha curcas] |
| 15 |
Hb_000120_370 |
0.0958718478 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |
| 16 |
Hb_000110_190 |
0.0960489961 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 17 |
Hb_001014_210 |
0.0963364979 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 18 |
Hb_003086_170 |
0.0965903445 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 19 |
Hb_007558_100 |
0.0967334826 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000984_170 |
0.09715387 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |