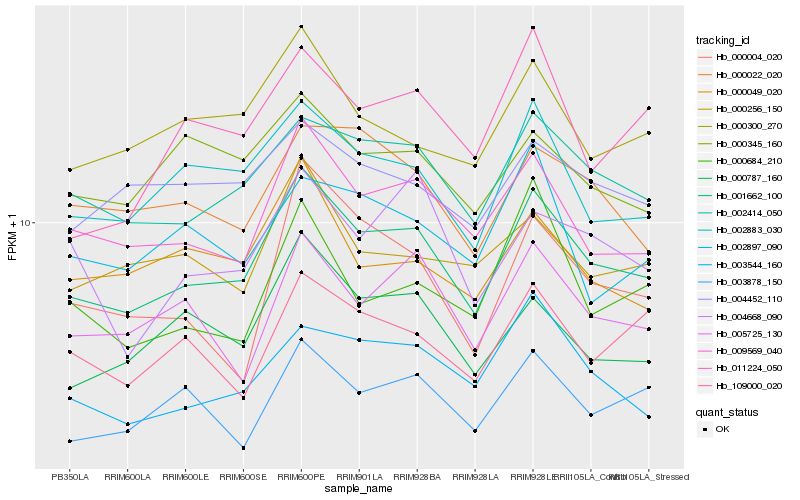
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001662_100 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 2 |
Hb_002414_050 |
0.0504155965 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 3 |
Hb_009569_040 |
0.0559567553 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 4 |
Hb_002883_030 |
0.0627626059 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 5 |
Hb_003544_160 |
0.0673115758 |
- |
- |
- |
| 6 |
Hb_000787_160 |
0.0692115988 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003878_150 |
0.069696041 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 8 |
Hb_005725_130 |
0.0720448645 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 9 |
Hb_000300_270 |
0.0743106054 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_000022_020 |
0.075993882 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 11 |
Hb_109000_020 |
0.0768774995 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 12 |
Hb_004452_110 |
0.0771516723 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000256_150 |
0.0783195715 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 14 |
Hb_000049_020 |
0.0784475744 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 15 |
Hb_000345_160 |
0.0800876347 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 16 |
Hb_000684_210 |
0.0839566582 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 17 |
Hb_002897_090 |
0.0844613301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004668_090 |
0.0849957154 |
- |
- |
PREDICTED: uncharacterized protein LOC105630440 [Jatropha curcas] |
| 19 |
Hb_011224_050 |
0.085486563 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Populus euphratica] |
| 20 |
Hb_000004_020 |
0.0855664309 |
- |
- |
conserved hypothetical protein [Ricinus communis] |