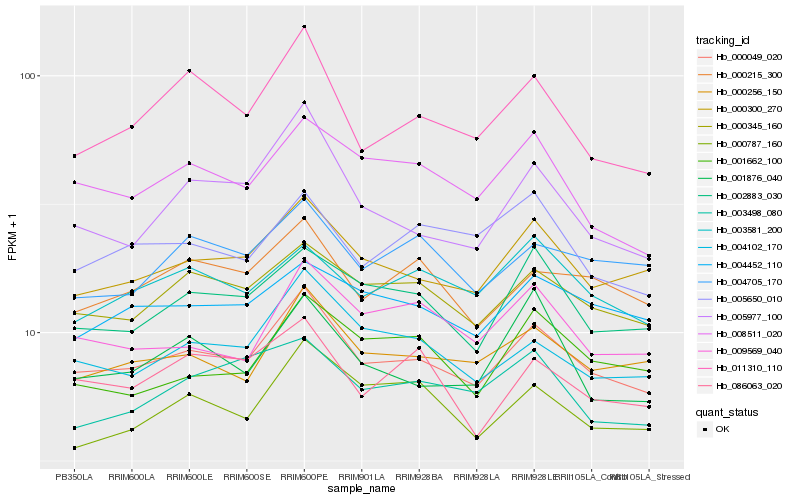
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000049_020 |
0.0 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 2 |
Hb_000300_270 |
0.0506830757 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_000345_160 |
0.053692412 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 4 |
Hb_000256_150 |
0.0554533317 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 5 |
Hb_004102_170 |
0.0611180699 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 6 |
Hb_002883_030 |
0.0658001126 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 7 |
Hb_000215_300 |
0.0674480404 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 8 |
Hb_004452_110 |
0.0676611824 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 9 |
Hb_008511_020 |
0.0689610401 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 10 |
Hb_003581_200 |
0.070259373 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 11 |
Hb_011310_110 |
0.0707734087 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009569_040 |
0.0716849948 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 13 |
Hb_000787_160 |
0.0726339298 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004705_170 |
0.0752192677 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 15 |
Hb_005977_100 |
0.0752617186 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 16 |
Hb_086063_020 |
0.0758536062 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 17 |
Hb_005650_010 |
0.0763864627 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_001876_040 |
0.0782896384 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 19 |
Hb_003498_080 |
0.0784314525 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 20 |
Hb_001662_100 |
0.0784475744 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |