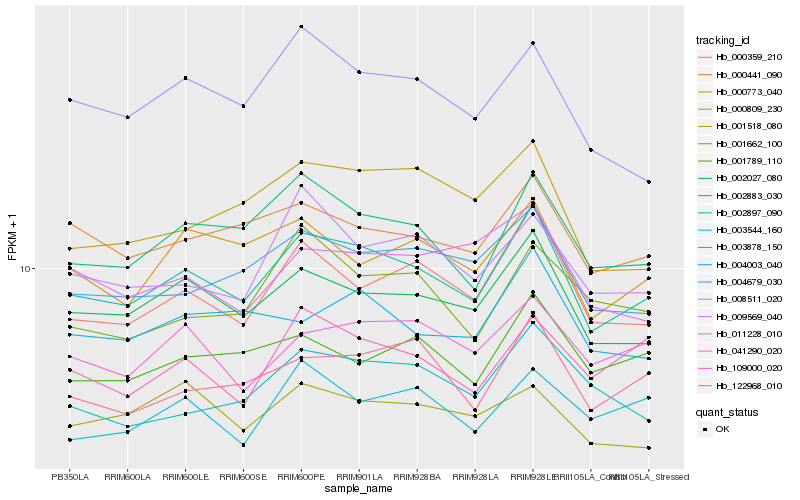
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002897_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002027_080 |
0.0509366312 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 3 |
Hb_002883_030 |
0.0592105994 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 4 |
Hb_000359_210 |
0.0606076846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004679_030 |
0.0676749788 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 6 |
Hb_000441_090 |
0.0693850689 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 7 |
Hb_122968_010 |
0.0711737409 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 8 |
Hb_000773_040 |
0.0738278538 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 9 |
Hb_008511_020 |
0.0746713869 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 10 |
Hb_041290_020 |
0.0765889118 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 11 |
Hb_009569_040 |
0.0776237552 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 12 |
Hb_004003_040 |
0.0792621123 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 13 |
Hb_001789_110 |
0.0793103904 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_000809_230 |
0.0793230883 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |
| 15 |
Hb_109000_020 |
0.0810011304 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 16 |
Hb_003544_160 |
0.082002122 |
- |
- |
- |
| 17 |
Hb_001518_080 |
0.0830198593 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 18 |
Hb_011228_010 |
0.0832504477 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_003878_150 |
0.0838959033 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 20 |
Hb_001662_100 |
0.0844613301 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |