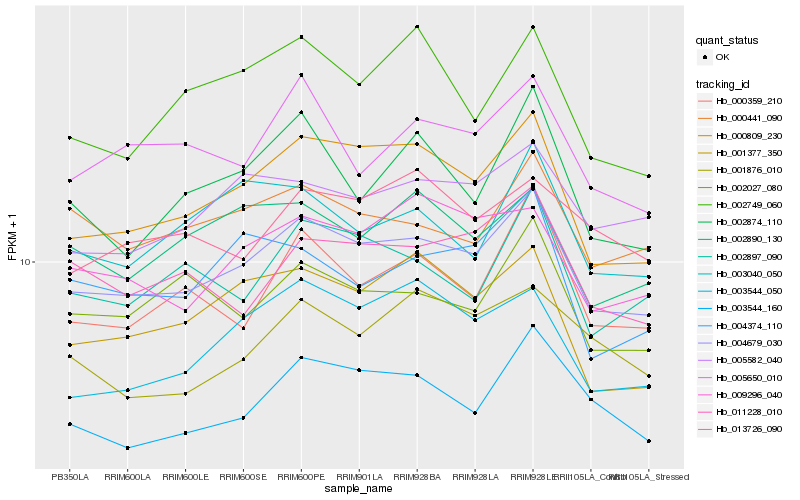
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004679_030 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 2 |
Hb_000809_230 |
0.0417933499 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003544_160 |
0.0615834616 |
- |
- |
- |
| 4 |
Hb_009296_040 |
0.0641324051 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 5 |
Hb_001377_350 |
0.0645100287 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 6 |
Hb_005582_040 |
0.0649380901 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 7 |
Hb_011228_010 |
0.0659437147 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000359_210 |
0.0669182444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002027_080 |
0.0670007988 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 10 |
Hb_000441_090 |
0.0671765657 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 11 |
Hb_002890_130 |
0.0675671168 |
- |
- |
tip120, putative [Ricinus communis] |
| 12 |
Hb_002897_090 |
0.0676749788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003544_050 |
0.0684246981 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005650_010 |
0.0688921164 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_002874_110 |
0.0721064711 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 16 |
Hb_004374_110 |
0.0734834619 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 17 |
Hb_002749_060 |
0.0736870053 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 18 |
Hb_003040_050 |
0.073906181 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 19 |
Hb_013726_090 |
0.0745419115 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 20 |
Hb_001876_010 |
0.0755377611 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |