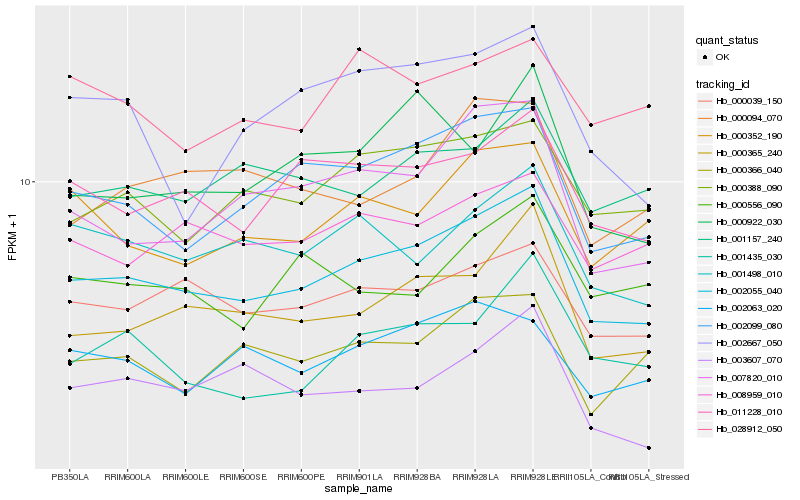
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002055_040 |
0.0 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 2 |
Hb_002099_080 |
0.0639706376 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 3 |
Hb_000365_240 |
0.066301461 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 4 |
Hb_011228_010 |
0.0705367808 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000039_150 |
0.071333119 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007820_010 |
0.0749354632 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: protein FLOWERING LOCUS D [Jatropha curcas] |
| 7 |
Hb_000388_090 |
0.075210663 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_001498_010 |
0.0794811333 |
- |
- |
PREDICTED: uncharacterized protein LOC105648994 [Jatropha curcas] |
| 9 |
Hb_000352_190 |
0.0809360353 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 41 [Jatropha curcas] |
| 10 |
Hb_008959_010 |
0.0822943623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001157_240 |
0.0839999351 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 12 |
Hb_002063_020 |
0.0852889499 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17140 [Jatropha curcas] |
| 13 |
Hb_000366_040 |
0.0865558556 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 14 |
Hb_000922_030 |
0.0868559439 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000094_070 |
0.0870604032 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001435_030 |
0.0910465621 |
- |
- |
PREDICTED: uncharacterized protein LOC105645073 [Jatropha curcas] |
| 17 |
Hb_002667_050 |
0.0935440953 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 18 |
Hb_003607_070 |
0.0942632258 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910 [Jatropha curcas] |
| 19 |
Hb_028912_050 |
0.0962813263 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000556_090 |
0.0963547769 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |