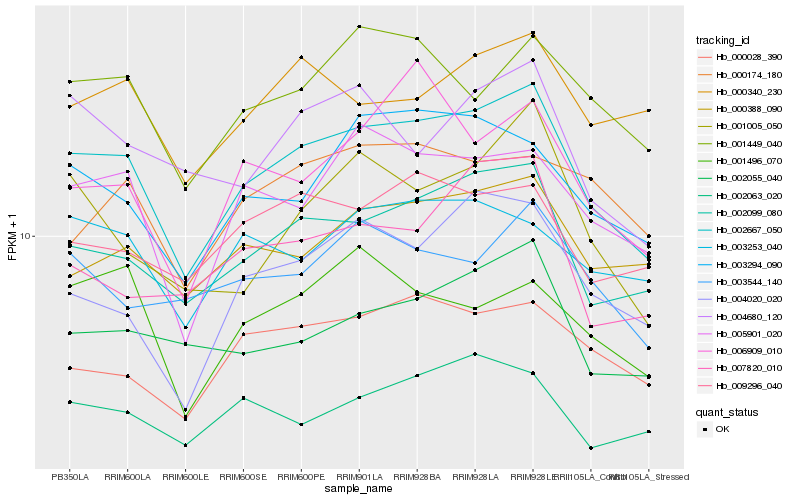
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002667_050 |
0.0 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 2 |
Hb_002099_080 |
0.0649629923 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 3 |
Hb_005901_020 |
0.0843080891 |
- |
- |
PREDICTED: elongation factor Tu GTP-binding domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_003294_090 |
0.0872144663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004020_020 |
0.0888924015 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g65820 [Jatropha curcas] |
| 6 |
Hb_001449_040 |
0.0909112346 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000388_090 |
0.0920570076 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_000028_390 |
0.0921597058 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 9 |
Hb_002055_040 |
0.0935440953 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 10 |
Hb_009296_040 |
0.0944194883 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 11 |
Hb_003253_040 |
0.096298269 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 12 |
Hb_002063_020 |
0.0973977606 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17140 [Jatropha curcas] |
| 13 |
Hb_001005_050 |
0.098849013 |
- |
- |
hypothetical protein Csa_3G599480 [Cucumis sativus] |
| 14 |
Hb_004680_120 |
0.0989180501 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 15 |
Hb_003544_140 |
0.1020251143 |
- |
- |
PREDICTED: protein GDAP2 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_000340_230 |
0.1023019141 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 17 |
Hb_001496_070 |
0.1028057154 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 18 |
Hb_007820_010 |
0.1034491793 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: protein FLOWERING LOCUS D [Jatropha curcas] |
| 19 |
Hb_006909_010 |
0.103599981 |
- |
- |
5-oxoprolinase, putative [Ricinus communis] |
| 20 |
Hb_000174_180 |
0.1050729668 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |