| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
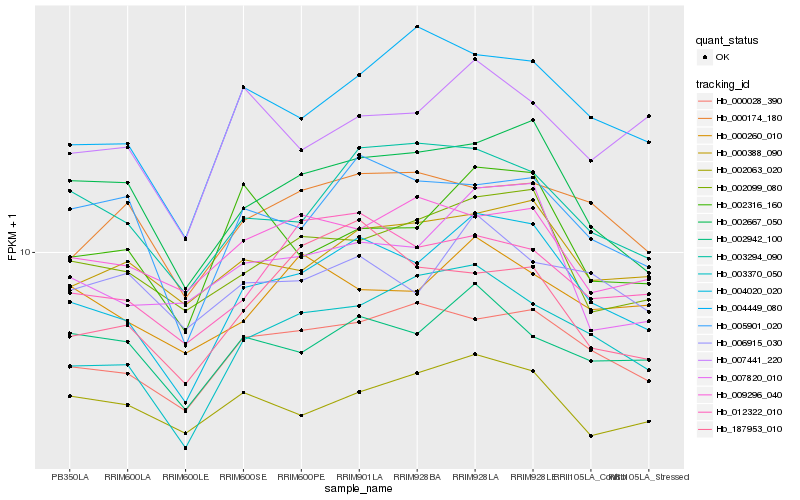
Hb_004020_020 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g65820 [Jatropha curcas] |
| 2 |
Hb_003370_050 |
0.0799439185 |
- |
- |
PREDICTED: uncharacterized protein LOC103697550 [Phoenix dactylifera] |
| 3 |
Hb_002667_050 |
0.0888924015 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 4 |
Hb_000028_390 |
0.0921902385 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 5 |
Hb_002099_080 |
0.0929841488 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 6 |
Hb_003294_090 |
0.0954995888 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007820_010 |
0.0999369815 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: protein FLOWERING LOCUS D [Jatropha curcas] |
| 8 |
Hb_004449_080 |
0.100532015 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 9 |
Hb_000388_090 |
0.1011143678 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_005901_020 |
0.1041974132 |
- |
- |
PREDICTED: elongation factor Tu GTP-binding domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_002316_160 |
0.105112668 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 12 |
Hb_012322_010 |
0.1077910071 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 13 |
Hb_002942_100 |
0.108639453 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61990, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000174_180 |
0.1152945644 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_002063_020 |
0.1153018088 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17140 [Jatropha curcas] |
| 16 |
Hb_006915_030 |
0.1174879664 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 8 [Jatropha curcas] |
| 17 |
Hb_007441_220 |
0.1188137102 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 18 |
Hb_000260_010 |
0.1193581893 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 1-like [Jatropha curcas] |
| 19 |
Hb_187953_010 |
0.1197574008 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_009296_040 |
0.1205891688 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |