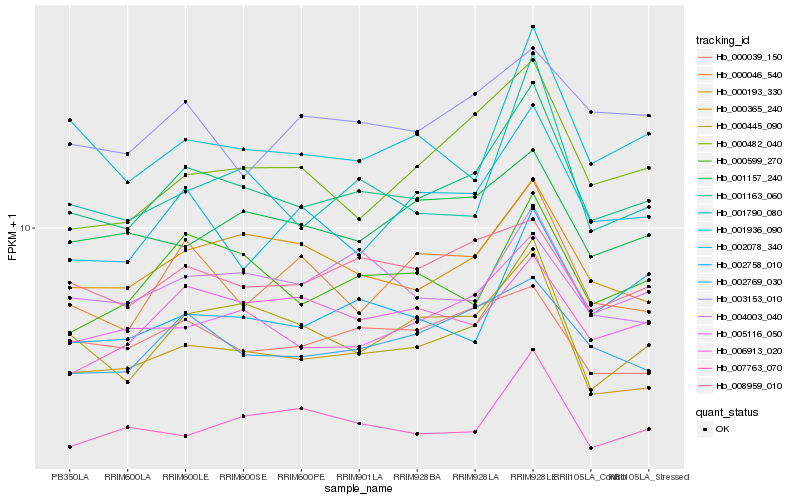
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001163_060 |
0.0 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 2 |
Hb_000445_090 |
0.0669121156 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 3 |
Hb_000193_330 |
0.0699314699 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 4 |
Hb_002769_030 |
0.0706646398 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000365_240 |
0.0751530023 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 6 |
Hb_000599_270 |
0.0756997656 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta isoform X3 [Jatropha curcas] |
| 7 |
Hb_004003_040 |
0.0769165131 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 8 |
Hb_001790_080 |
0.0772830687 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 9 |
Hb_008959_010 |
0.0782954932 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007763_070 |
0.0795385459 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001936_090 |
0.0806171664 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_005116_050 |
0.0812554028 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_000482_040 |
0.0819076586 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 14 |
Hb_000046_540 |
0.0827842579 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 15 |
Hb_001157_240 |
0.0834764479 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 16 |
Hb_002078_340 |
0.0852438897 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000039_150 |
0.086322576 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003153_010 |
0.0875883315 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 19 |
Hb_006913_020 |
0.0876418355 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002758_010 |
0.0878379581 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |