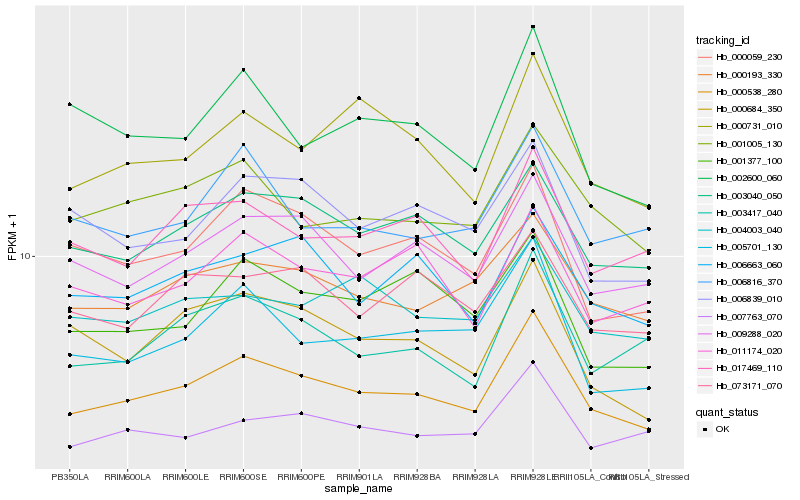
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000538_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 2 |
Hb_003040_050 |
0.0657767038 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 3 |
Hb_000193_330 |
0.0679690173 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 4 |
Hb_009288_020 |
0.0697359716 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 5 |
Hb_001005_130 |
0.072808823 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 6 |
Hb_000059_230 |
0.0747699072 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 7 |
Hb_005701_130 |
0.0761508424 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 8 |
Hb_003417_040 |
0.0763301059 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 9 |
Hb_007763_070 |
0.0772230751 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 10 |
Hb_006663_060 |
0.0777370565 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 11 |
Hb_017469_110 |
0.0791726714 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 12 |
Hb_000731_010 |
0.0795486013 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_011174_020 |
0.083930147 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 14 |
Hb_004003_040 |
0.0840016026 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_073171_070 |
0.0840202312 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 16 |
Hb_006816_370 |
0.0845495252 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 17 |
Hb_006839_010 |
0.085226876 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 18 |
Hb_001377_100 |
0.0860569226 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000684_350 |
0.0870113425 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002600_060 |
0.088966729 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |