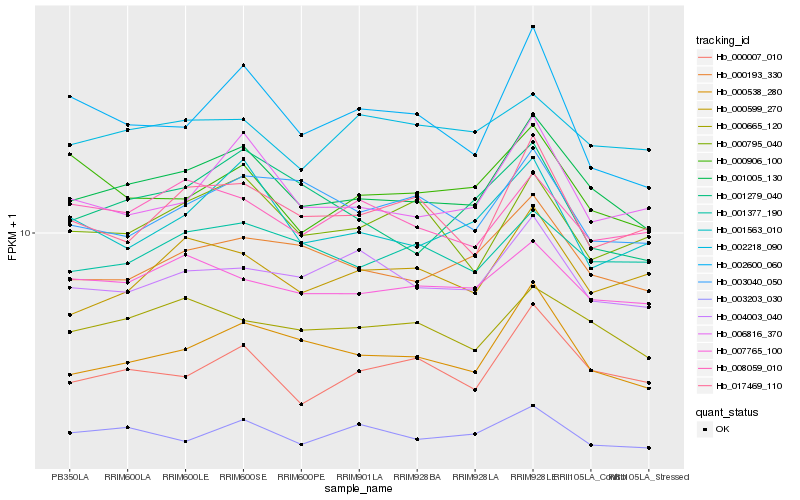
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001005_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 2 |
Hb_000193_330 |
0.0712768114 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 3 |
Hb_000538_280 |
0.072808823 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 4 |
Hb_006816_370 |
0.074521385 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 5 |
Hb_000665_120 |
0.0798943909 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 6 |
Hb_000007_010 |
0.0800539479 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001377_190 |
0.0830375369 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007765_100 |
0.0848438413 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 9 |
Hb_004003_040 |
0.0870792878 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 10 |
Hb_001563_010 |
0.0888411924 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 11 |
Hb_017469_110 |
0.0889337664 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 12 |
Hb_000599_270 |
0.0905131732 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta isoform X3 [Jatropha curcas] |
| 13 |
Hb_000906_100 |
0.0932553773 |
- |
- |
PREDICTED: uncharacterized protein LOC105648070 [Jatropha curcas] |
| 14 |
Hb_001279_040 |
0.0934535134 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 15 |
Hb_000795_040 |
0.0940563772 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 16 |
Hb_008059_010 |
0.0951787576 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003040_050 |
0.0956035098 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 18 |
Hb_002218_090 |
0.0963333564 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 19 |
Hb_003203_030 |
0.0964839349 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 20 |
Hb_002600_060 |
0.0966735402 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |