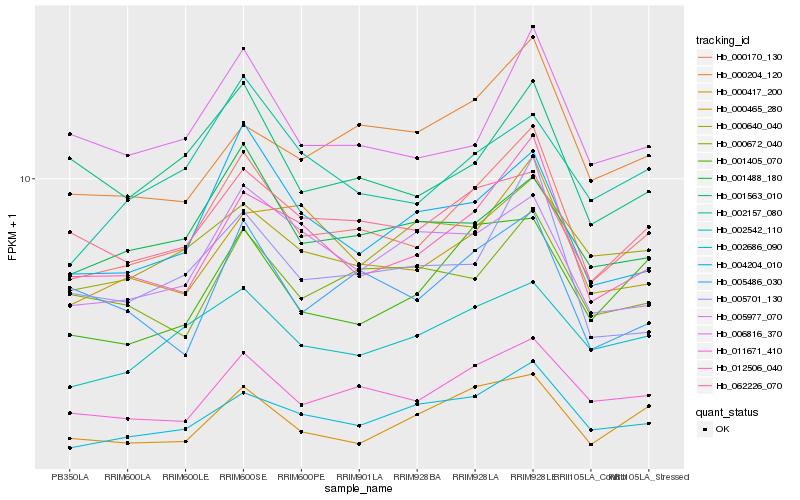
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000170_130 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 2 |
Hb_011671_410 |
0.0634619083 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 3 |
Hb_012506_040 |
0.0685338678 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 4 |
Hb_002686_090 |
0.0744198169 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 5 |
Hb_002542_110 |
0.0822605782 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 6 |
Hb_006816_370 |
0.0837279939 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 7 |
Hb_062226_070 |
0.0857472306 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004204_010 |
0.086270758 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 9 |
Hb_001405_070 |
0.0884168808 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 10 |
Hb_005701_130 |
0.0886797684 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 11 |
Hb_001488_180 |
0.0887351395 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001563_010 |
0.089064189 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 13 |
Hb_000204_120 |
0.0894243512 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 14 |
Hb_005486_030 |
0.0908474878 |
transcription factor |
TF Family: PHD |
hypothetical protein POPTR_0017s10890g [Populus trichocarpa] |
| 15 |
Hb_000465_280 |
0.0911846423 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 16 |
Hb_000672_040 |
0.0924738916 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 17 |
Hb_000417_200 |
0.0926542423 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002157_080 |
0.0948950388 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005977_070 |
0.0956510959 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000640_040 |
0.0962206287 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |