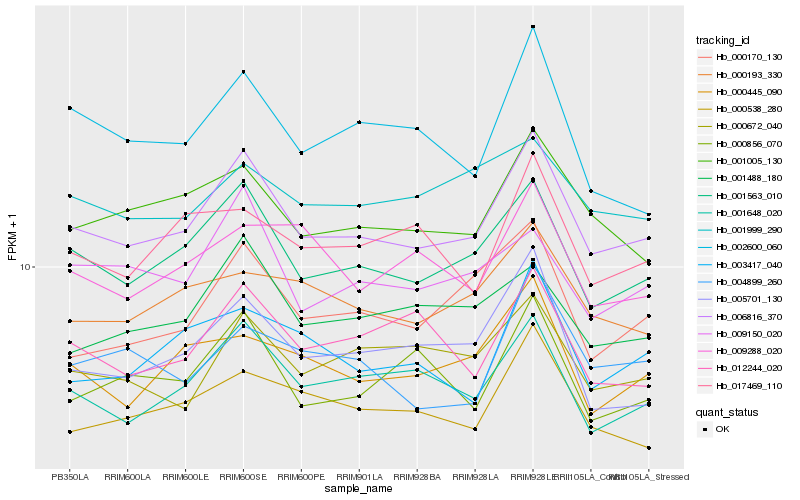
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_370 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 2 |
Hb_001563_010 |
0.0456018194 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 3 |
Hb_001648_020 |
0.0715136437 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 4 |
Hb_001005_130 |
0.074521385 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 5 |
Hb_000170_130 |
0.0837279939 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 6 |
Hb_000538_280 |
0.0845495252 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 7 |
Hb_000193_330 |
0.0854975502 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 8 |
Hb_000672_040 |
0.085756797 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 9 |
Hb_001488_180 |
0.0859539739 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000445_090 |
0.0863451346 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 11 |
Hb_005701_130 |
0.0867232681 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 12 |
Hb_009150_020 |
0.0880070148 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 13 |
Hb_000856_070 |
0.0897521041 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 14 |
Hb_002600_060 |
0.0900686882 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 15 |
Hb_012244_020 |
0.0900970153 |
- |
- |
calpain, putative [Ricinus communis] |
| 16 |
Hb_003417_040 |
0.0910553019 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 17 |
Hb_017469_110 |
0.0936785737 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 18 |
Hb_004899_260 |
0.0942111004 |
- |
- |
PREDICTED: uncharacterized protein LOC105638006 [Jatropha curcas] |
| 19 |
Hb_001999_290 |
0.0950956599 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_009288_020 |
0.0953228748 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |