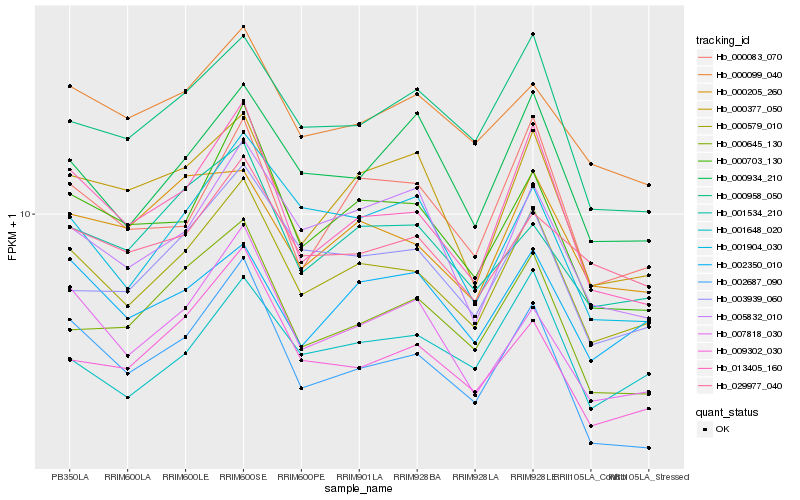
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000579_010 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 2 |
Hb_013405_160 |
0.0572223689 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000703_130 |
0.0624887192 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 4 |
Hb_003939_060 |
0.0691825146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000958_050 |
0.0693589403 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 6 |
Hb_000083_070 |
0.0698192813 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000099_040 |
0.0727834925 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_000934_210 |
0.0728362732 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 9 |
Hb_000645_130 |
0.0736330745 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 10 |
Hb_000377_050 |
0.0740766787 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 11 |
Hb_007818_030 |
0.0741212622 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 12 |
Hb_029977_040 |
0.0744158217 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 13 |
Hb_001534_210 |
0.074666156 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001648_020 |
0.0748781245 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 15 |
Hb_000205_260 |
0.0752475883 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 16 |
Hb_009302_030 |
0.075525838 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 17 |
Hb_005832_010 |
0.0755427317 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002350_010 |
0.0756976217 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 19 |
Hb_002687_090 |
0.0761625759 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 20 |
Hb_001904_030 |
0.0763751212 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |