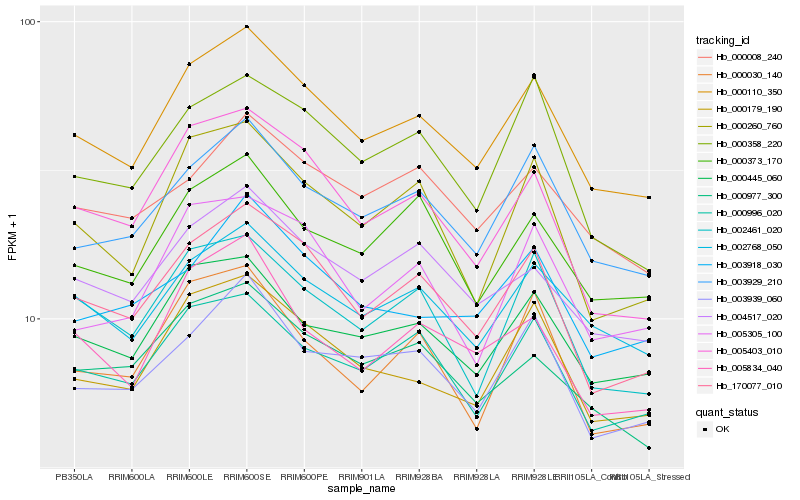
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_350 |
0.0 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 2 |
Hb_170077_010 |
0.0436766908 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000179_190 |
0.0453304745 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 4 |
Hb_003929_210 |
0.0482613833 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 5 |
Hb_002768_050 |
0.0506135435 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 6 |
Hb_000445_060 |
0.0506645769 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000996_020 |
0.0548239259 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 8 |
Hb_004517_020 |
0.0554104619 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 9 |
Hb_003939_060 |
0.0561138516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000373_170 |
0.060505237 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 11 |
Hb_005403_010 |
0.06265649 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 12 |
Hb_005834_040 |
0.0637057051 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 13 |
Hb_000977_300 |
0.0643061947 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 14 |
Hb_000358_220 |
0.06452043 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 15 |
Hb_002461_020 |
0.0647829107 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000260_760 |
0.0648852866 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 17 |
Hb_003918_030 |
0.0656342118 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000008_240 |
0.0664799341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000030_140 |
0.0680505224 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005305_100 |
0.0686575027 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |