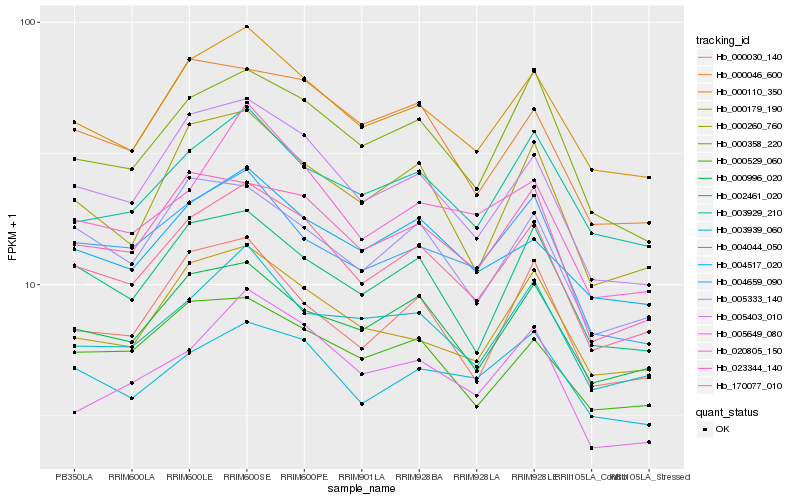
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_170077_010 |
0.0 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000110_350 |
0.0436766908 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 3 |
Hb_005403_010 |
0.0472023044 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 4 |
Hb_023344_140 |
0.0561733921 |
- |
- |
PREDICTED: WD repeat-containing protein 70 [Jatropha curcas] |
| 5 |
Hb_000358_220 |
0.0586502636 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 6 |
Hb_000996_020 |
0.0614530139 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 7 |
Hb_002461_020 |
0.0627423849 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000260_760 |
0.0635168127 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 9 |
Hb_004659_090 |
0.0659044319 |
- |
- |
PREDICTED: uncharacterized protein LOC105629935 [Jatropha curcas] |
| 10 |
Hb_003939_060 |
0.0659171192 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004517_020 |
0.06633335 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 12 |
Hb_000179_190 |
0.067279683 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 13 |
Hb_005333_140 |
0.0682916838 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003929_210 |
0.0687262507 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 15 |
Hb_000046_600 |
0.0694684934 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 16 |
Hb_000529_060 |
0.0694962943 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005649_080 |
0.0695129266 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 18 |
Hb_000030_140 |
0.069853639 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_020805_150 |
0.0699289125 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_004044_050 |
0.0700372013 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |