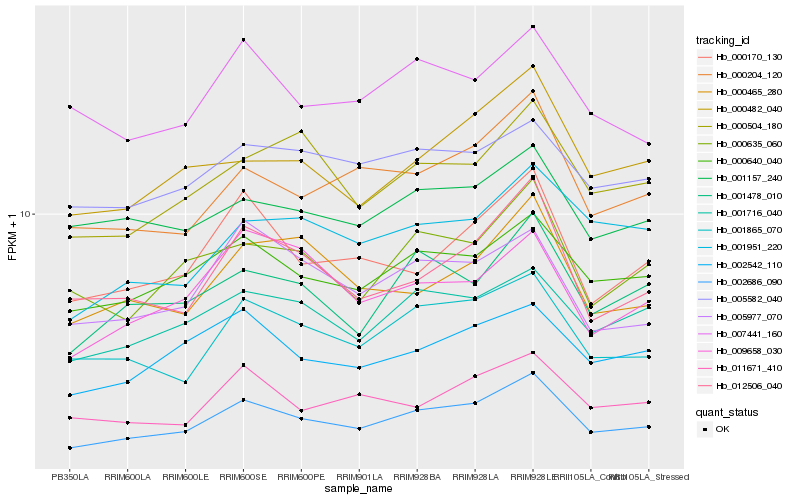
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_090 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 2 |
Hb_000640_040 |
0.0639075702 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001865_070 |
0.0654555002 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000204_120 |
0.0692407027 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 5 |
Hb_001716_040 |
0.0703138393 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 6 |
Hb_002542_110 |
0.0738168629 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 7 |
Hb_000170_130 |
0.0744198169 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 8 |
Hb_001478_010 |
0.0745102346 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48250, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000635_060 |
0.0749982651 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 10 |
Hb_012506_040 |
0.0751726303 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 11 |
Hb_000482_040 |
0.0754005412 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 12 |
Hb_000504_180 |
0.0756347012 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 13 |
Hb_001951_220 |
0.0806822416 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 14 |
Hb_011671_410 |
0.0817842597 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 15 |
Hb_000465_280 |
0.0826529871 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 16 |
Hb_005977_070 |
0.0833671965 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005582_040 |
0.0835183591 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 18 |
Hb_001157_240 |
0.086576162 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 19 |
Hb_009658_030 |
0.0884308794 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 20 |
Hb_007441_160 |
0.0887392224 |
- |
- |
RNA binding protein, putative [Ricinus communis] |