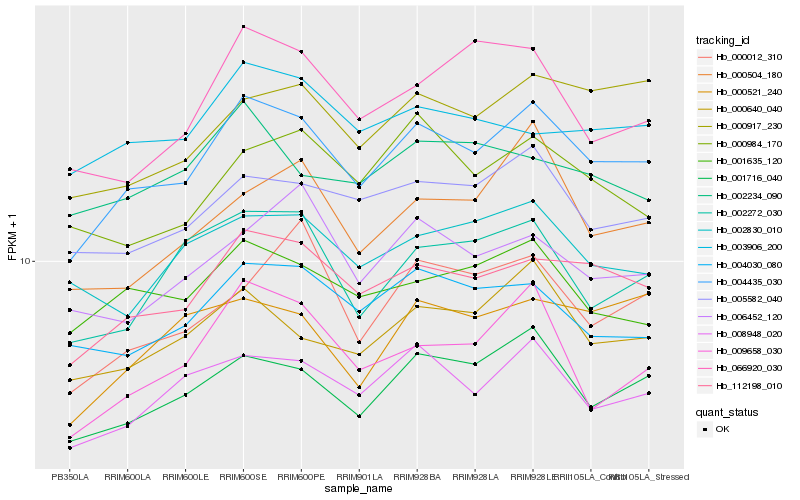
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004435_030 |
0.0 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 2 |
Hb_112198_010 |
0.0458169641 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 3 |
Hb_001716_040 |
0.0579061576 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 4 |
Hb_008948_020 |
0.0611139166 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 5 |
Hb_000640_040 |
0.0701535101 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005582_040 |
0.0704187077 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 7 |
Hb_001635_120 |
0.0737096767 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 8 |
Hb_004030_080 |
0.0743944658 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 9 |
Hb_009658_030 |
0.0762448802 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 10 |
Hb_000917_230 |
0.0780022646 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Jatropha curcas] |
| 11 |
Hb_002272_030 |
0.0780525199 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 12 |
Hb_000504_180 |
0.0809872721 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 13 |
Hb_003906_200 |
0.0811748113 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 14 |
Hb_002830_010 |
0.0818269645 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_006452_120 |
0.0819630078 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 16 |
Hb_000984_170 |
0.0823903126 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 17 |
Hb_000521_240 |
0.0828823574 |
- |
- |
PREDICTED: uncharacterized protein LOC105650339 isoform X2 [Jatropha curcas] |
| 18 |
Hb_066920_030 |
0.083547763 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 19 |
Hb_000012_310 |
0.0856607464 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_002234_090 |
0.0866895297 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |