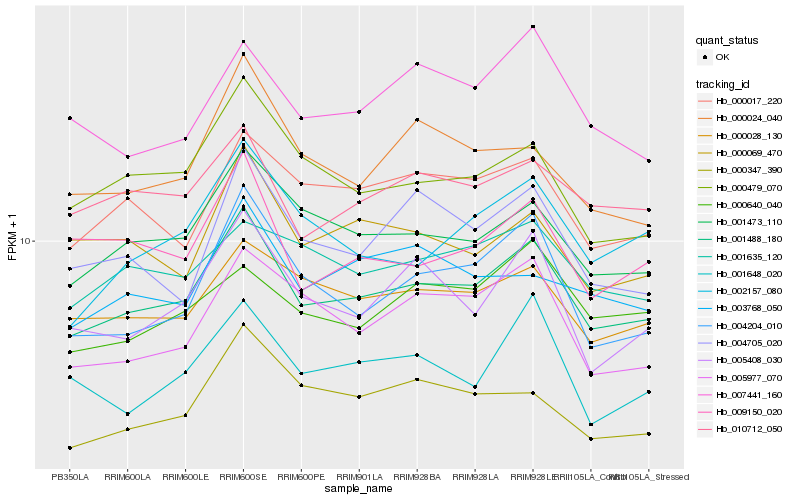
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001473_110 |
0.0565005432 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 3 |
Hb_000028_130 |
0.0653364759 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000479_070 |
0.0674648078 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 5 |
Hb_004204_010 |
0.0687582043 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 6 |
Hb_010712_050 |
0.0695594211 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 7 |
Hb_000017_220 |
0.0700163295 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 8 |
Hb_005408_030 |
0.0703289172 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 9 |
Hb_005977_070 |
0.0735296759 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000347_390 |
0.0763218012 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 11 |
Hb_001635_120 |
0.0765002007 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 12 |
Hb_002157_080 |
0.0766813892 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 13 |
Hb_009150_020 |
0.076772561 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 14 |
Hb_004705_020 |
0.0773053734 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 15 |
Hb_000069_470 |
0.0774896396 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001648_020 |
0.0780528323 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 17 |
Hb_000024_040 |
0.0781974101 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 18 |
Hb_003768_050 |
0.0784789161 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007441_160 |
0.0788448739 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000640_040 |
0.0789574483 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |