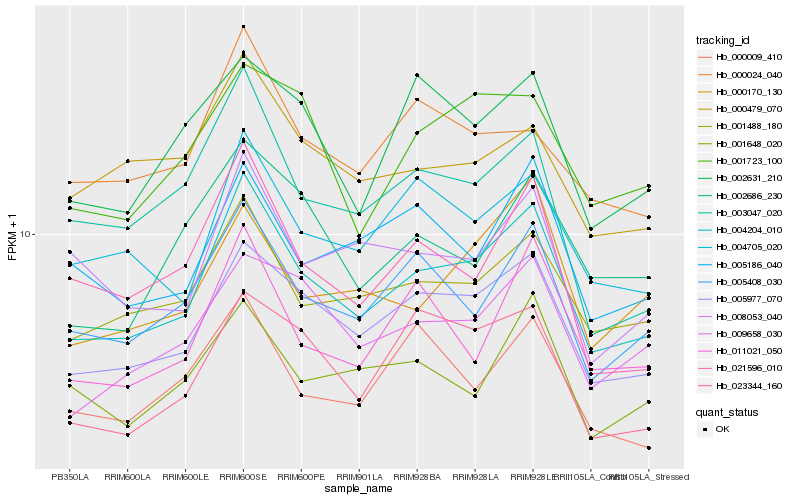
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004204_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 2 |
Hb_005977_070 |
0.0503941401 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001488_180 |
0.0687582043 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005408_030 |
0.0701254669 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 5 |
Hb_004705_020 |
0.0788144225 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 6 |
Hb_009658_030 |
0.0810792252 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 7 |
Hb_011021_050 |
0.0823838113 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 8 |
Hb_021596_010 |
0.0834829289 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 9 |
Hb_023344_160 |
0.0838613732 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 10 |
Hb_005186_040 |
0.0859996748 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000170_130 |
0.086270758 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 12 |
Hb_008053_040 |
0.0876995367 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 13 |
Hb_000009_410 |
0.0883129026 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 14 |
Hb_003047_020 |
0.0887399092 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_001648_020 |
0.0897030148 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 16 |
Hb_000024_040 |
0.090244126 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 17 |
Hb_001723_100 |
0.0909560071 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002686_230 |
0.0914992728 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_000479_070 |
0.0917417477 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 20 |
Hb_002631_210 |
0.0919765539 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |