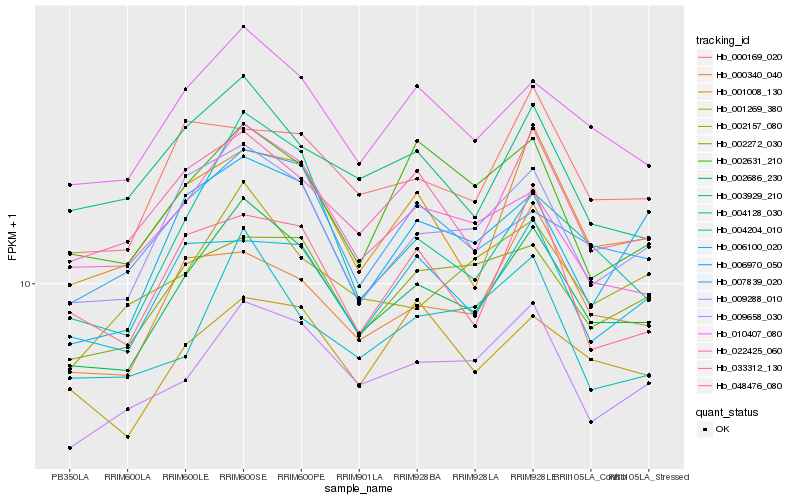
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_230 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_010407_080 |
0.0592060798 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009288_010 |
0.062206291 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_009658_030 |
0.0721901069 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 5 |
Hb_033312_130 |
0.0736213376 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001008_130 |
0.0773849668 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 7 |
Hb_022425_060 |
0.0775654004 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 8 |
Hb_007839_020 |
0.0787287266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004128_030 |
0.082727008 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 8-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_006100_020 |
0.0848728375 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 11 |
Hb_002272_030 |
0.0852130252 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 12 |
Hb_003929_210 |
0.085461551 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 13 |
Hb_006970_050 |
0.087127467 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 14 |
Hb_000169_020 |
0.0872035402 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 15 |
Hb_000340_040 |
0.0886263562 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 16 |
Hb_002631_210 |
0.0894670597 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 17 |
Hb_002157_080 |
0.0894819153 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001269_380 |
0.0905897499 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004204_010 |
0.0914992728 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 20 |
Hb_048476_080 |
0.0917936176 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |