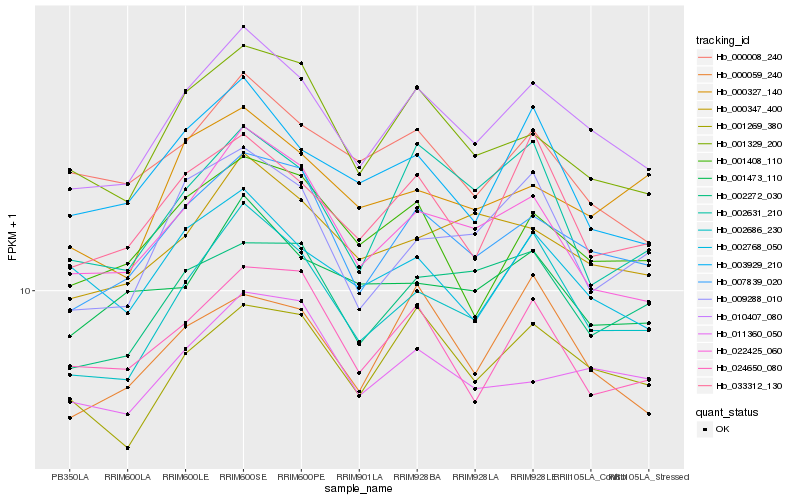
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010407_080 |
0.0 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007839_020 |
0.0457663079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002686_230 |
0.0592060798 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_022425_060 |
0.0669051519 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_001269_380 |
0.0679256664 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001329_200 |
0.0682226301 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 7 |
Hb_033312_130 |
0.0735626155 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000347_400 |
0.0753573456 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001408_110 |
0.0760103764 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 10 |
Hb_011360_050 |
0.0782843123 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003929_210 |
0.0791714679 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 12 |
Hb_000008_240 |
0.0811650453 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002768_050 |
0.0818586045 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 14 |
Hb_009288_010 |
0.0827539424 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_002272_030 |
0.0841271026 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 16 |
Hb_000059_240 |
0.0844442627 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 17 |
Hb_024650_080 |
0.0847838908 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 18 |
Hb_001473_110 |
0.0851012904 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 19 |
Hb_000327_140 |
0.0865871735 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002631_210 |
0.0873457391 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |