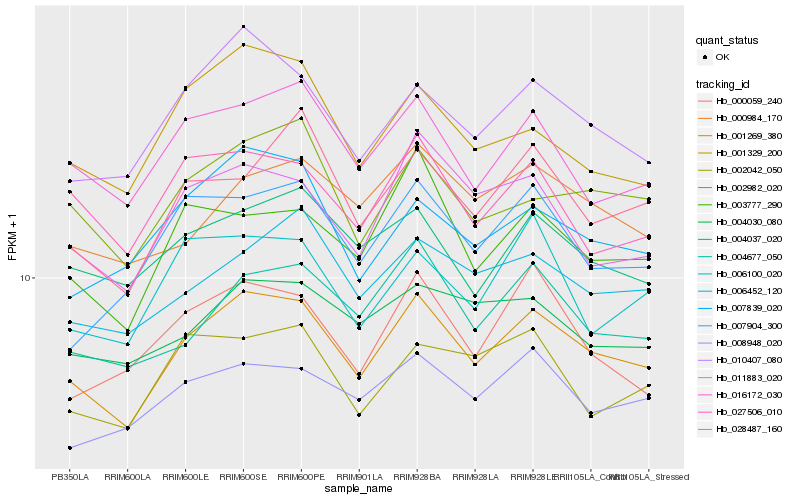
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_380 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 2 |
Hb_010407_080 |
0.0679256664 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004677_050 |
0.0732031053 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_016172_030 |
0.0739707706 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 5 |
Hb_002982_020 |
0.0747970209 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 6 |
Hb_006452_120 |
0.0762694489 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 7 |
Hb_008948_020 |
0.0764043339 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 8 |
Hb_011883_020 |
0.0767001866 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 9 |
Hb_004037_020 |
0.0771546373 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003777_290 |
0.0778079925 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 11 |
Hb_028487_160 |
0.0781336167 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 12 |
Hb_007839_020 |
0.0800600934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000984_170 |
0.0806411441 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 14 |
Hb_004030_080 |
0.0819427564 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 15 |
Hb_027506_010 |
0.0838134749 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 16 |
Hb_001329_200 |
0.0840042765 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 17 |
Hb_000059_240 |
0.0841366695 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 18 |
Hb_007904_300 |
0.0850984897 |
- |
- |
copine, putative [Ricinus communis] |
| 19 |
Hb_006100_020 |
0.0851027386 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 20 |
Hb_002042_050 |
0.0863026506 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |