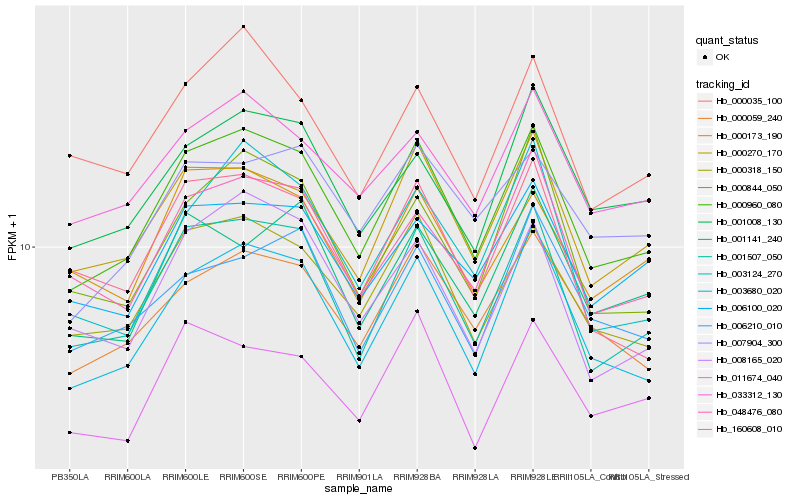
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000960_080 |
0.0 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001507_050 |
0.0402619327 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_003680_020 |
0.0415168041 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000318_150 |
0.0470032813 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 5 |
Hb_000270_170 |
0.0565735047 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 6 |
Hb_000059_240 |
0.0612391525 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 7 |
Hb_033312_130 |
0.0626059796 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 8 |
Hb_048476_080 |
0.0644085063 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 9 |
Hb_008165_020 |
0.0671350878 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001008_130 |
0.0684394338 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 11 |
Hb_006100_020 |
0.0686966777 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 12 |
Hb_000173_190 |
0.0695906513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000844_050 |
0.0714525111 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 14 |
Hb_011674_040 |
0.0730953003 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000035_100 |
0.0745219153 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_007904_300 |
0.0752595962 |
- |
- |
copine, putative [Ricinus communis] |
| 17 |
Hb_003124_270 |
0.0763743673 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 18 |
Hb_006210_010 |
0.0765458919 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 19 |
Hb_160608_010 |
0.0765847819 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 20 |
Hb_001141_240 |
0.0766380071 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |